You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003512_01659
You are here: Home > Sequence: MGYG000003512_01659
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA11524 sp900769075 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; UBA11524; UBA11524 sp900769075 | |||||||||||
| CAZyme ID | MGYG000003512_01659 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6468; End: 7631 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 65 | 344 | 5.6e-89 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 9.47e-54 | 67 | 347 | 18 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.92e-22 | 36 | 377 | 46 | 394 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG2723 | BglB | 0.005 | 67 | 184 | 57 | 172 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADK66823.1 | 3.84e-88 | 24 | 376 | 23 | 365 |
| QPB75694.1 | 1.03e-85 | 23 | 383 | 29 | 393 |
| ADU21113.1 | 1.09e-85 | 3 | 361 | 6 | 359 |
| BAA92146.1 | 1.09e-85 | 3 | 361 | 6 | 359 |
| QPZ86391.1 | 1.13e-83 | 32 | 361 | 1 | 325 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XSU_A | 1.11e-91 | 30 | 359 | 9 | 333 | GH5-4broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens],6XSU_B GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens] |
| 6Q1I_A | 2.05e-74 | 33 | 380 | 13 | 356 | GH5-4broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum],6Q1I_B GH5-4 broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum] |
| 3AYR_A | 7.84e-73 | 32 | 376 | 18 | 359 | GH5endoglucanase EglA from a ruminal fungus [Piromyces rhizinflatus] |
| 3AYS_A | 6.16e-72 | 32 | 376 | 18 | 359 | GH5endoglucanase from a ruminal fungus in complex with cellotriose [Piromyces rhizinflatus] |
| 4NF7_A | 2.91e-69 | 30 | 375 | 4 | 362 | Crystalstructure of the GH5 family catalytic domain of Endo-1,4-beta-glucanase Cel5C from Butyrivibrio proteoclasticus. [Butyrivibrio proteoclasticus B316] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23661 | 2.57e-76 | 26 | 379 | 63 | 402 | Endoglucanase B OS=Ruminococcus albus OX=1264 GN=celB PE=3 SV=1 |
| P16216 | 7.34e-75 | 26 | 373 | 61 | 394 | Endoglucanase 1 OS=Ruminococcus albus OX=1264 GN=Eg I PE=1 SV=1 |
| Q12647 | 1.13e-74 | 26 | 376 | 17 | 362 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| P54937 | 1.02e-72 | 33 | 380 | 38 | 381 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P23660 | 2.94e-69 | 32 | 377 | 25 | 363 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
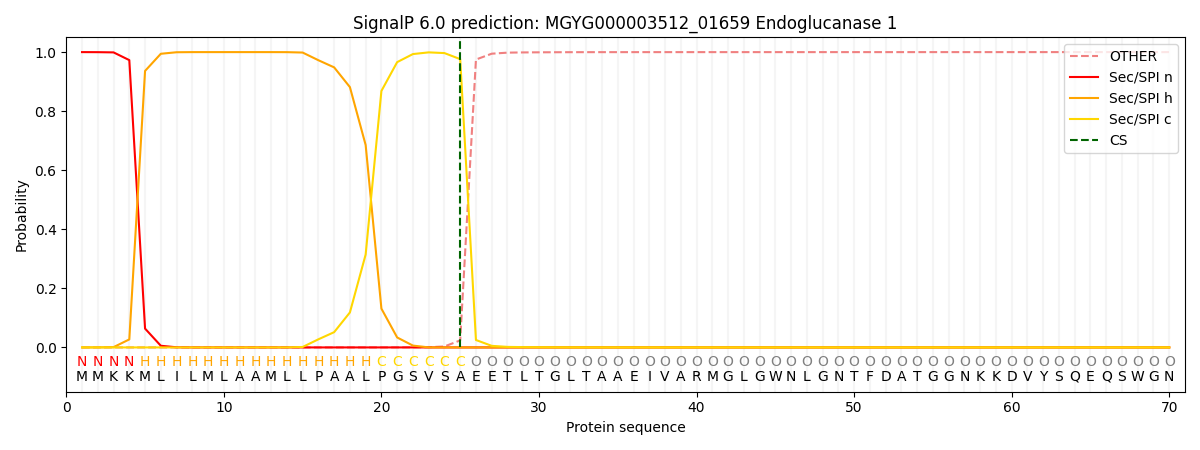
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000319 | 0.998846 | 0.000206 | 0.000217 | 0.000203 | 0.000180 |
