You are browsing environment: HUMAN GUT
MGYG000003512_01157
Basic Information
help
Species
UBA11524 sp900769075
Lineage
Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; CAG-74; UBA11524; UBA11524 sp900769075
CAZyme ID
MGYG000003512_01157
CAZy Family
GH28
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
488
54090.17
4.617
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003512
2777574
MAG
Fiji
Oceania
Gene Location
Start: 1883;
End: 3349
Strand: -
No EC number prediction in MGYG000003512_01157.
Family
Start
End
Evalue
family coverage
GH28
192
467
4.6e-42
0.7630769230769231
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG5434
Pgu1
3.00e-12
188
320
178
336
Polygalacturonase [Carbohydrate transport and metabolism].
more
pfam00295
Glyco_hydro_28
2.43e-04
199
470
48
310
Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism.
more
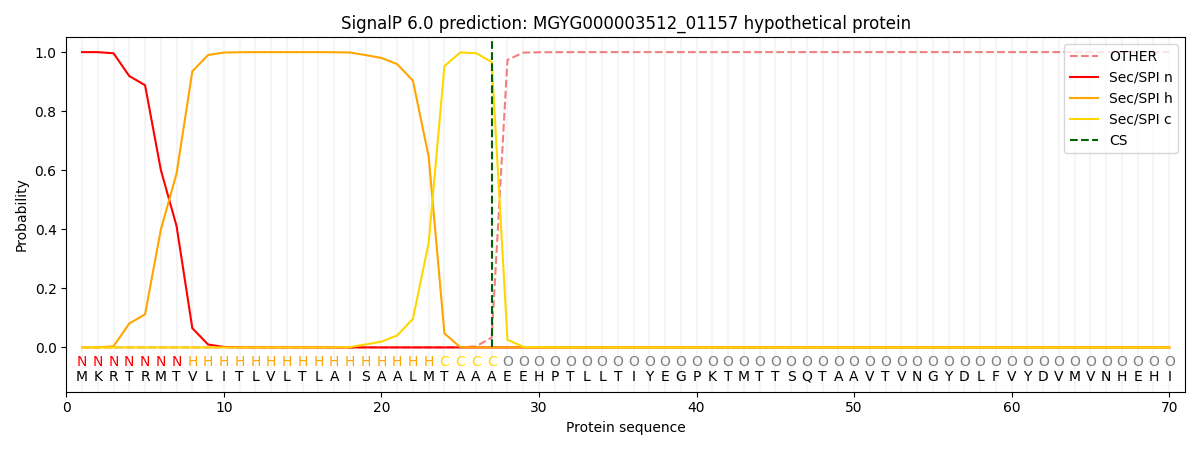
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000428
0.998723
0.000231
0.000258
0.000176
0.000155