You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003489_00404
You are here: Home > Sequence: MGYG000003489_00404
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotellamassilia sp900768625 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotellamassilia; Prevotellamassilia sp900768625 | |||||||||||
| CAZyme ID | MGYG000003489_00404 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1474; End: 4455 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 346 | 701 | 4.5e-44 | 0.9269005847953217 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 2.33e-60 | 340 | 711 | 1 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| COG4409 | NanH | 3.01e-04 | 331 | 482 | 251 | 425 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFQ12979.1 | 4.28e-111 | 329 | 721 | 11 | 398 |
| QUB76590.1 | 1.64e-110 | 25 | 983 | 57 | 1044 |
| QUB85277.1 | 2.20e-110 | 25 | 971 | 38 | 1011 |
| ALO48969.1 | 3.08e-110 | 329 | 724 | 16 | 403 |
| QYR10380.1 | 9.75e-109 | 322 | 720 | 10 | 395 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7LBU_A | 2.29e-17 | 341 | 697 | 82 | 415 | ChainA, Exo-alpha-sialidase [Cutibacterium acnes],7LBV_A Chain A, Exo-alpha-sialidase [Cutibacterium acnes] |
| 1EUR_A | 6.17e-16 | 339 | 705 | 11 | 353 | Sialidase[Micromonospora viridifaciens],1EUS_A Sialidase Complexed With 2-Deoxy-2,3-Dehydro-N- Acetylneuraminic Acid [Micromonospora viridifaciens] |
| 1EUT_A | 2.09e-15 | 339 | 705 | 11 | 353 | Sialidase,Large 68kd Form, Complexed With Galactose [Micromonospora viridifaciens],1EUU_A Sialidase Or Neuraminidase, Large 68kd Form [Micromonospora viridifaciens] |
| 1WCQ_A | 6.27e-15 | 339 | 705 | 7 | 349 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
| 1W8N_A | 1.09e-14 | 339 | 705 | 7 | 349 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 1.24e-14 | 339 | 705 | 53 | 395 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P31206 | 1.91e-06 | 351 | 571 | 201 | 419 | Sialidase OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=nanH PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
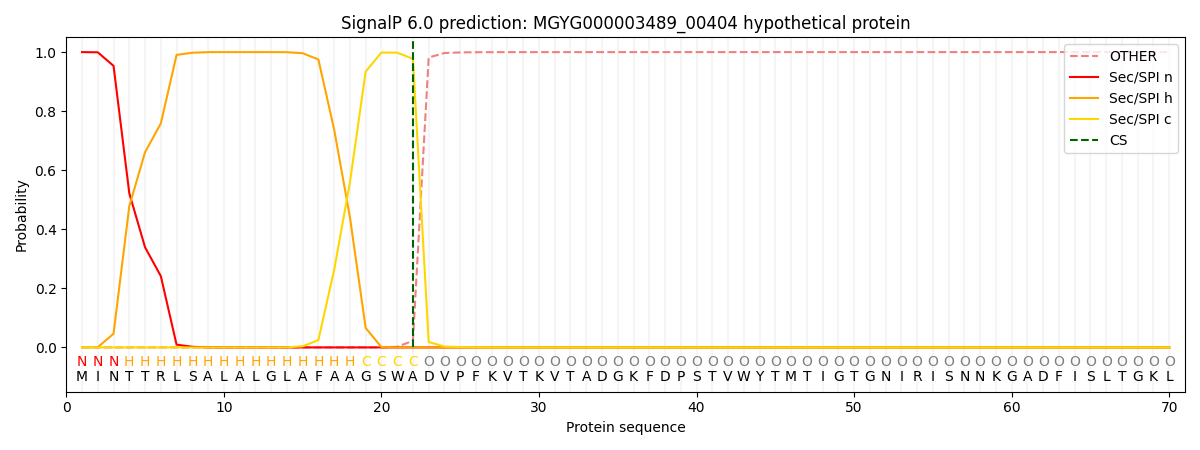
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000286 | 0.999034 | 0.000174 | 0.000184 | 0.000169 | 0.000151 |
