You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003434_01221
You are here: Home > Sequence: MGYG000003434_01221
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-279; | |||||||||||
| CAZyme ID | MGYG000003434_01221 | |||||||||||
| CAZy Family | GH144 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2125; End: 3708 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH144 | 121 | 524 | 1.1e-164 | 0.9876237623762376 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam10091 | Glycoamylase | 3.06e-91 | 291 | 512 | 1 | 214 | Putative glucoamylase. The structure of UniProt:Q5LIB7 has an alpha/alpha toroid fold and is similar structurally to a number of glucoamylases. Most of these structural homologs are glucoamylases, involved in breaking down complex sugars (e.g. starch). The biologically relevant state is likely to be monomeric. The putative active site is located at the centre of the toroid with a well defined large cavity. |
| COG5368 | COG5368 | 6.20e-90 | 122 | 523 | 9 | 429 | Uncharacterized protein [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADQ80137.1 | 2.31e-161 | 49 | 525 | 42 | 523 |
| QXV65763.1 | 1.23e-157 | 121 | 525 | 45 | 445 |
| SMD46157.1 | 3.54e-157 | 121 | 525 | 252 | 651 |
| QDH81568.1 | 7.76e-156 | 113 | 525 | 243 | 650 |
| QGA23454.1 | 2.93e-155 | 80 | 525 | 81 | 530 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4GL3_A | 1.42e-143 | 121 | 525 | 13 | 416 | Crystalstructure of a putative glucoamylase (BACUNI_03963) from Bacteroides uniformis ATCC 8492 at 2.01 A resolution [Bacteroides uniformis ATCC 8492] |
| 5GZH_A | 2.61e-134 | 121 | 525 | 24 | 436 | Endo-beta-1,2-glucanasefrom Chitinophaga pinensis - ligand free form [Chitinophaga pinensis DSM 2588],5GZH_B Endo-beta-1,2-glucanase from Chitinophaga pinensis - ligand free form [Chitinophaga pinensis DSM 2588] |
| 5GZK_A | 2.73e-134 | 94 | 525 | 19 | 456 | Endo-beta-1,2-glucanasefrom Chitinophaga pinensis - sophorotriose and glucose complex [Chitinophaga pinensis DSM 2588],5GZK_B Endo-beta-1,2-glucanase from Chitinophaga pinensis - sophorotriose and glucose complex [Chitinophaga pinensis DSM 2588] |
| 3EU8_A | 3.95e-134 | 113 | 525 | 8 | 427 | Crystalstructure of putative glucoamylase (YP_210071.1) from Bacteroides fragilis NCTC 9343 at 2.12 A resolution [Bacteroides fragilis NCTC 9343],3EU8_B Crystal structure of putative glucoamylase (YP_210071.1) from Bacteroides fragilis NCTC 9343 at 2.12 A resolution [Bacteroides fragilis NCTC 9343],3EU8_C Crystal structure of putative glucoamylase (YP_210071.1) from Bacteroides fragilis NCTC 9343 at 2.12 A resolution [Bacteroides fragilis NCTC 9343],3EU8_D Crystal structure of putative glucoamylase (YP_210071.1) from Bacteroides fragilis NCTC 9343 at 2.12 A resolution [Bacteroides fragilis NCTC 9343] |
| 4QT9_A | 1.10e-130 | 121 | 525 | 19 | 430 | Crystalstructure of a putative glucoamylase (BACCAC_03554) from Bacteroides caccae ATCC 43185 at 2.05 A resolution [Bacteroides caccae ATCC 43185],4QT9_B Crystal structure of a putative glucoamylase (BACCAC_03554) from Bacteroides caccae ATCC 43185 at 2.05 A resolution [Bacteroides caccae ATCC 43185] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A6LGF6 | 5.81e-111 | 121 | 522 | 299 | 717 | Exo beta-1,2-glucooligosaccharide sophorohydrolase (non-reducing end) OS=Parabacteroides distasonis (strain ATCC 8503 / DSM 20701 / CIP 104284 / JCM 5825 / NCTC 11152) OX=435591 GN=BDI_3064 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
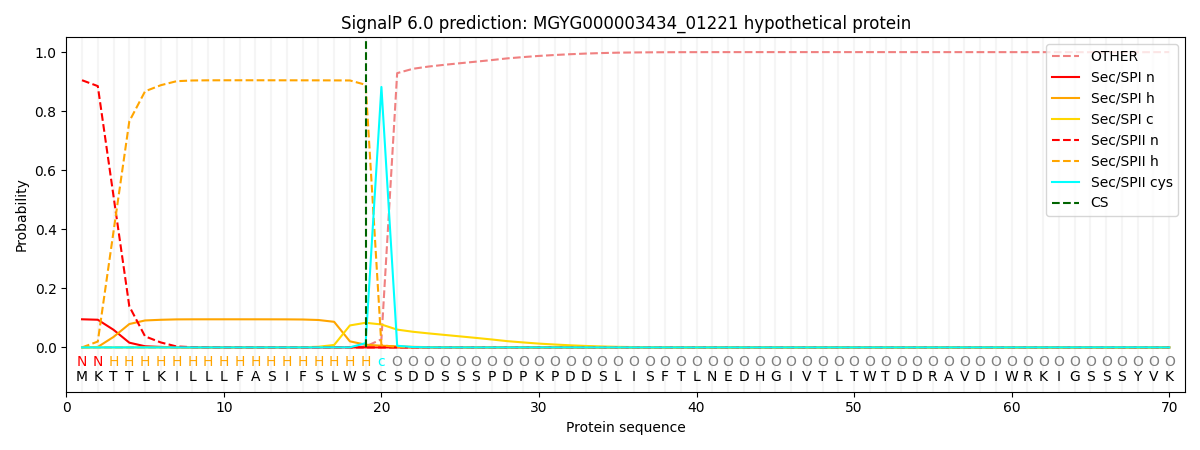
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000123 | 0.093170 | 0.906591 | 0.000037 | 0.000049 | 0.000039 |
