You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003420_00649
You are here: Home > Sequence: MGYG000003420_00649
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
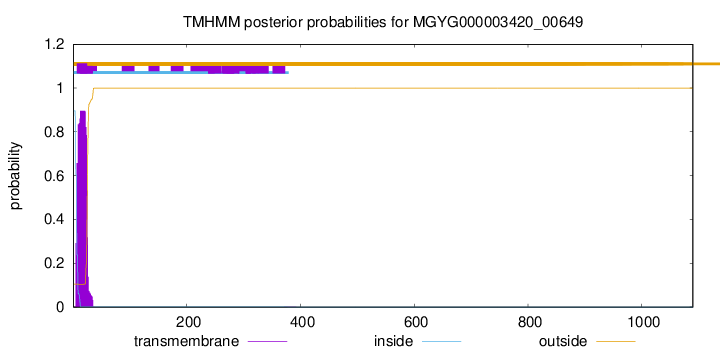
TMHMM annotations
Basic Information help
| Species | UBA4372 sp900766785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA4372; UBA4372 sp900766785 | |||||||||||
| CAZyme ID | MGYG000003420_00649 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3183; End: 6455 Strand: + | |||||||||||
Full Sequence Download help
| MKNRNIPRYA FASLLLLMVT VVSSARNKYL FVYFTGNEPQ QEQIRYAVSD DGFHYTPLNG | 60 |
| GDPVVASDSI ALTGCVRDPH ILRTQDGEYL MVATDMRSSL GWSSNRGIVL MKSRDLIHWQ | 120 |
| HHTVHFPKRY EGKNFARVTR VWAPQTIYDE QAGKYMVYFS LLTDDGTIPY DRVYWAYANA | 180 |
| DFTDLEGDPQ VLFDFHAPAI DTDIVRDHTG LYHLFFKTEQ EGAHKGIRQY VFQDLHAPQE | 240 |
| WTLLDGFCEQ TKDNVEGAGV FPLIGGDWCL MYDCYTKGHY QFTRSSDLRT FTHVTDTKTE | 300 |
| GAFTPRHGTV IQISSQECRK LEQSLAKRRA KMMQNTFVAG DGTFLLHGEP FVVKAAEIHY | 360 |
| PRIPRPYWEH RIQMCKALGM NTICIYIFWN IHEQQEGVFD FTGQNDVAEF CRLAQKNGMY | 420 |
| VIVRPGPYVC AEWEMGGLPW WLLKKKDVRL REPDEFFMAA VDRFEQKVAE QLAPLTIQQG | 480 |
| GPIIMVQVEN EYGSYGENKQ YIAQIRDTLR KYWDKDGSKP TTLFQCDWNS NFEKNGLDDL | 540 |
| VWTMNFGTGA NIDDQFRRLR QLRPHTPLMC SEFWSGWFDK WGARHETRPA QDMVNGIDEM | 600 |
| LSKGISFSLY MTHGGTSFGH WAGANSPGFA PDVTSYDYDA PINEYGCATE KYHLLRQTLQ | 660 |
| KYSSQKLPAI PAAPARLISI PRMTLSLVSS LCMGVDSVAA SREPITFEEM NMGYGSMIYR | 720 |
| TDLPQIATGS TLHIDGHDFV QAFINGKYVG KVDRVKNERT LQLPPTQQGD RLTLLVEAMG | 780 |
| RINFGRAIKD FKGLIGDVSL TADVDGDEVT WTLKDWQMAR IKDSYSHALR ALSAPQSDMG | 840 |
| PLVDLPKPIG YYRTTFRLKQ TGDTFLNMET WGKGLVYLNG HALGRFWSIG PQQTLYCPGC | 900 |
| WLKKGENEII VIDVVGPREP VLWGQDKPEL DKLQLERSLR HNNIGDKPDL NSATPVAQGA | 960 |
| TKAGNGWQTI TFSQAAQGRF LAIQCSTTHD GKPVAVAEIY LKDKNGKRIP RDQWSVKYAN | 1020 |
| SENLQGNHTG DKAFDLQEST FWQTEQDASA PHLLVIDLGG EYTVTALEYL PRMEQGAPGS | 1080 |
| IKDYQIFILK | 1090 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 344 | 660 | 1.4e-116 | 0.993485342019544 |
| GH43 | 74 | 310 | 9.2e-76 | 0.9914163090128756 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01301 | Glyco_hydro_35 | 2.64e-154 | 343 | 657 | 1 | 313 | Glycosyl hydrolases family 35. |
| cd08983 | GH43_Bt3655-like | 2.98e-73 | 57 | 323 | 1 | 260 | Glycosyl hydrolase family 43 protein such as Bacteroides thetaiotaomicron VPI-5482 arabinofuranosidase Bt3655. This glycosyl hydrolase family 43 (GH43)-like family includes the characterized arabinofuranosidases (EC 3.2.1.55): Bacteroides thetaiotaomicron VPI-5482 (Bt3655;BT_3655) and Penicillium chrysogenum 31B Abf43B, as well as Bifidobacterium adolescentis ATCC 15703 beta-xylosidase (EC 3.2.1.37) BAD_1527. It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 includes enzymes with beta-xylosidase (EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanases (beta-xylanases) and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| PLN03059 | PLN03059 | 5.07e-56 | 343 | 916 | 36 | 715 | beta-galactosidase; Provisional |
| COG1874 | GanA | 3.44e-42 | 342 | 917 | 6 | 593 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| cd08978 | GH_F | 2.29e-11 | 77 | 313 | 1 | 239 | Glycosyl hydrolase families 43 and 62 form CAZY clan GH-F. This glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) includes family 43 (GH43) and 62 (GH62). GH43 includes enzymes with beta-xylosidase (EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanases (beta-xylanases) and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. GH62 includes enzymes characterized as arabinofuranosidases (alpha-L-arabinofuranosidases; EC 3.2.1.55) that specifically cleave either alpha-1,2 or alpha-1,3-L-arabinofuranose side chains from xylans. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many of the enzymes in this family display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. GH62 are also predicted to be inverting enzymes. A common structural feature of both, GH43 and GH62 enzymes, is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGB27610.1 | 0.0 | 337 | 1088 | 32 | 796 |
| QVJ81893.1 | 0.0 | 337 | 1088 | 24 | 786 |
| ADE83089.1 | 0.0 | 337 | 1088 | 24 | 786 |
| BCS84536.1 | 0.0 | 337 | 1088 | 30 | 778 |
| VEH15027.1 | 0.0 | 324 | 1088 | 17 | 783 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6EON_A | 1.03e-293 | 334 | 1088 | 25 | 772 | GalactanaseBT0290 [Bacteroides thetaiotaomicron VPI-5482] |
| 3D3A_A | 3.37e-237 | 332 | 946 | 3 | 605 | Crystalstructure of a beta-galactosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4MAD_A | 5.06e-128 | 341 | 914 | 21 | 578 | ChainA, Beta-galactosidase [Niallia circulans],4MAD_B Chain B, Beta-galactosidase [Niallia circulans] |
| 3WF3_A | 4.03e-113 | 343 | 930 | 41 | 634 | Crystalstructure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_B Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_C Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_D Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF4_A Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_B Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_C Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_D Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens] |
| 3THC_A | 4.06e-113 | 343 | 930 | 17 | 610 | Crystalstructure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_B Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_C Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_D Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THD_A Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_B Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_C Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_D Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16278 | 4.17e-112 | 343 | 930 | 40 | 633 | Beta-galactosidase OS=Homo sapiens OX=9606 GN=GLB1 PE=1 SV=2 |
| Q95LV1 | 6.02e-112 | 344 | 956 | 39 | 649 | Beta-galactosidase-1-like protein OS=Macaca fascicularis OX=9541 GN=GLB1L PE=2 SV=1 |
| Q60HF6 | 1.80e-111 | 344 | 930 | 41 | 633 | Beta-galactosidase OS=Macaca fascicularis OX=9541 GN=GLB1 PE=2 SV=1 |
| Q6UWU2 | 2.27e-111 | 340 | 956 | 35 | 649 | Beta-galactosidase-1-like protein OS=Homo sapiens OX=9606 GN=GLB1L PE=2 SV=1 |
| P48982 | 2.54e-110 | 336 | 896 | 29 | 572 | Beta-galactosidase OS=Xanthomonas manihotis OX=43353 GN=bga PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.260979 | 0.677977 | 0.059980 | 0.000365 | 0.000286 | 0.000388 |