You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003383_01079
You are here: Home > Sequence: MGYG000003383_01079
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
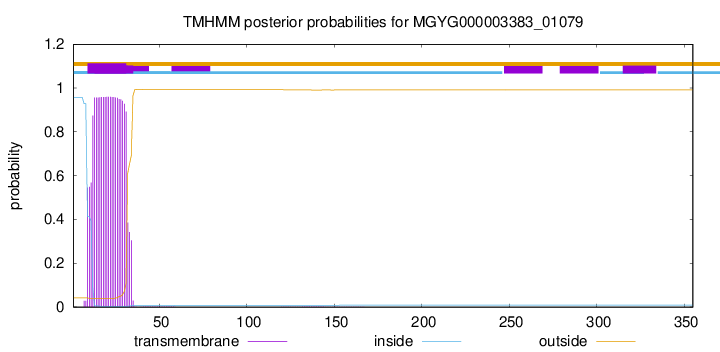
TMHMM annotations
Basic Information help
| Species | Bifidobacterium scardovii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium scardovii | |||||||||||
| CAZyme ID | MGYG000003383_01079 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 274275; End: 275342 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK08581 | PRK08581 | 5.46e-32 | 72 | 194 | 492 | 618 | amidase domain-containing protein. |
| COG3942 | COG3942 | 1.94e-21 | 84 | 182 | 68 | 158 | Surface antigen [Cell wall/membrane/envelope biogenesis]. |
| pfam05257 | CHAP | 3.82e-17 | 84 | 171 | 5 | 83 | CHAP domain. This domain corresponds to an amidase function. Many of these proteins are involved in cell wall metabolism of bacteria. This domain is found at the N-terminus of Escherichia coli gss, where it functions as a glutathionylspermidine amidase EC:3.5.1.78. This domain is found to be the catalytic domain of PlyCA. CHAP is the amidase domain of bifunctional Escherichia coli glutathionylspermidine synthetase/amidase, and it catalyzes the hydrolysis of Gsp (glutathionylspermidine) into glutathione and spermidine. |
| cd13925 | RPF | 0.001 | 287 | 328 | 10 | 53 | core lysozyme-like domain of resuscitation-promoting factor proteins. Resuscitation-promoting factor (RPF) proteins, found in various (G+C)-rich Gram-positive bacteria, act to reactivate cultures from stationary phase. This protein shares elements of the structural core of lysozyme and related proteins. Furthermore, it shares a conserved active site glutamate which is required for activity, and has a polysaccharide binding cleft that corresponds to the peptidoglycan binding cleft of lysozyme. Muralytic activity of Rpf in Micrococcus luteus correlates with resuscitation, supporting a mechanism dependent on cleavage of peptidoglycan by RPF. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUE01287.1 | 2.83e-214 | 1 | 355 | 1 | 356 |
| AHJ24910.1 | 5.02e-214 | 1 | 355 | 1 | 362 |
| AUE20777.1 | 5.02e-214 | 1 | 355 | 1 | 362 |
| AUE05354.1 | 5.02e-214 | 1 | 355 | 1 | 362 |
| ALE13022.1 | 7.92e-211 | 1 | 355 | 1 | 362 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T1Q_A | 2.40e-12 | 84 | 193 | 248 | 353 | ChainA, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_B Chain B, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_C Chain C, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_D Chain D, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 2LRJ_A | 2.47e-10 | 84 | 174 | 11 | 90 | ChainA, Staphyloxanthin biosynthesis protein, putative [Staphylococcus aureus subsp. aureus COL] |
| 4CGK_A | 2.92e-08 | 83 | 174 | 286 | 365 | Crystalstructure of the essential protein PcsB from Streptococcus pneumoniae [Streptococcus pneumoniae D39],4CGK_B Crystal structure of the essential protein PcsB from Streptococcus pneumoniae [Streptococcus pneumoniae D39] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q2G222 | 4.06e-11 | 84 | 193 | 508 | 613 | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=SAOUHSC_02979 PE=1 SV=1 |
| Q2G2J2 | 3.49e-09 | 74 | 191 | 156 | 263 | Staphylococcal secretory antigen ssaA2 OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=ssaA2 PE=1 SV=1 |
| Q5HDQ9 | 3.49e-09 | 74 | 191 | 156 | 263 | Staphylococcal secretory antigen ssaA2 OS=Staphylococcus aureus (strain COL) OX=93062 GN=ssaA2 PE=3 SV=1 |
| Q7A423 | 3.49e-09 | 74 | 191 | 156 | 263 | Staphylococcal secretory antigen ssaA2 OS=Staphylococcus aureus (strain N315) OX=158879 GN=ssaA2 PE=1 SV=1 |
| Q99RX4 | 3.49e-09 | 74 | 191 | 156 | 263 | Staphylococcal secretory antigen ssaA2 OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=ssaA2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000456 | 0.999570 | 0.000000 | 0.000003 | 0.000000 |