You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003360_01566
You are here: Home > Sequence: MGYG000003360_01566
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Enterobacter_D kobei_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Enterobacter_D; Enterobacter_D kobei_A | |||||||||||
| CAZyme ID | MGYG000003360_01566 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 117789; End: 119966 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06543 | GH18_PF-ChiA-like | 7.21e-112 | 275 | 581 | 1 | 294 | PF-ChiA is an uncharacterized chitinase found in the hyperthermophilic archaeon Pyrococcus furiosus with a glycosyl hydrolase family 18 (GH18) catalytic domain as well as a cellulose-binding domain. Members of this domain family are found not only in archaea but also in eukaryotes and prokaryotes. PF-ChiA exhibits hydrolytic activity toward both colloidal and crystalline (beta/alpha) chitins at high temperature. |
| NF035930 | lectin_2 | 5.84e-13 | 599 | 698 | 125 | 214 | lectin. Lectins are important adhesin proteins, which bind carbohydrate structures on host cell surface. The carbohydrate specificity of diverse lectins to a large extent dictates bacteria tissue tropism by mediating specific attachment to unique host sites expressing the corresponding carbohydrate receptor. |
| pfam00652 | Ricin_B_lectin | 1.62e-12 | 592 | 718 | 2 | 126 | Ricin-type beta-trefoil lectin domain. |
| NF035930 | lectin_2 | 8.79e-12 | 603 | 678 | 169 | 238 | lectin. Lectins are important adhesin proteins, which bind carbohydrate structures on host cell surface. The carbohydrate specificity of diverse lectins to a large extent dictates bacteria tissue tropism by mediating specific attachment to unique host sites expressing the corresponding carbohydrate receptor. |
| cd00161 | RICIN | 2.25e-09 | 599 | 720 | 8 | 124 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCU57561.1 | 0.0 | 1 | 725 | 1 | 725 |
| QLO82529.1 | 0.0 | 1 | 723 | 1 | 723 |
| QKE22293.1 | 0.0 | 1 | 723 | 1 | 723 |
| QYO51062.1 | 0.0 | 1 | 723 | 1 | 723 |
| QMR48447.1 | 0.0 | 1 | 584 | 1 | 584 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2DSK_A | 4.11e-42 | 266 | 583 | 1 | 292 | Crystalstructure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus [Pyrococcus furiosus],2DSK_B Crystal structure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus [Pyrococcus furiosus] |
| 3A4W_A | 1.99e-41 | 266 | 583 | 1 | 292 | Crystalstructures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3A4W_B Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus] |
| 3A4X_A | 5.12e-41 | 266 | 583 | 1 | 292 | Crystalstructures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3A4X_B Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3AFB_A Crystal structures of catalytic site mutants of active domain 2 of chitinase from Pyrococcus furiosus [Pyrococcus furiosus],3AFB_B Crystal structures of catalytic site mutants of active domain 2 of chitinase from Pyrococcus furiosus [Pyrococcus furiosus] |
| 6OGD_C | 1.12e-10 | 25 | 150 | 24 | 160 | Cryo-EMstructure of YenTcA in its prepore state [Yersinia entomophaga],6OGD_F Cryo-EM structure of YenTcA in its prepore state [Yersinia entomophaga],6OGD_I Cryo-EM structure of YenTcA in its prepore state [Yersinia entomophaga],6OGD_L Cryo-EM structure of YenTcA in its prepore state [Yersinia entomophaga],6OGD_O Cryo-EM structure of YenTcA in its prepore state [Yersinia entomophaga] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P13656 | 6.13e-264 | 21 | 584 | 333 | 895 | Probable bifunctional chitinase/lysozyme OS=Escherichia coli (strain K12) OX=83333 GN=chiA PE=1 SV=2 |
| O74199 | 5.90e-213 | 39 | 553 | 1 | 511 | Endochitinase 11 OS=Metarhizium anisopliae OX=5530 GN=chi11 PE=1 SV=1 |
| P14529 | 2.00e-14 | 278 | 553 | 44 | 299 | Chitinase OS=Saccharopolyspora erythraea (strain ATCC 11635 / DSM 40517 / JCM 4748 / NBRC 13426 / NCIMB 8594 / NRRL 2338) OX=405948 GN=chiA2 PE=1 SV=2 |
| B6A879 | 6.14e-10 | 25 | 150 | 24 | 160 | Chitinase 2 OS=Yersinia entomophaga OX=935293 GN=chi2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
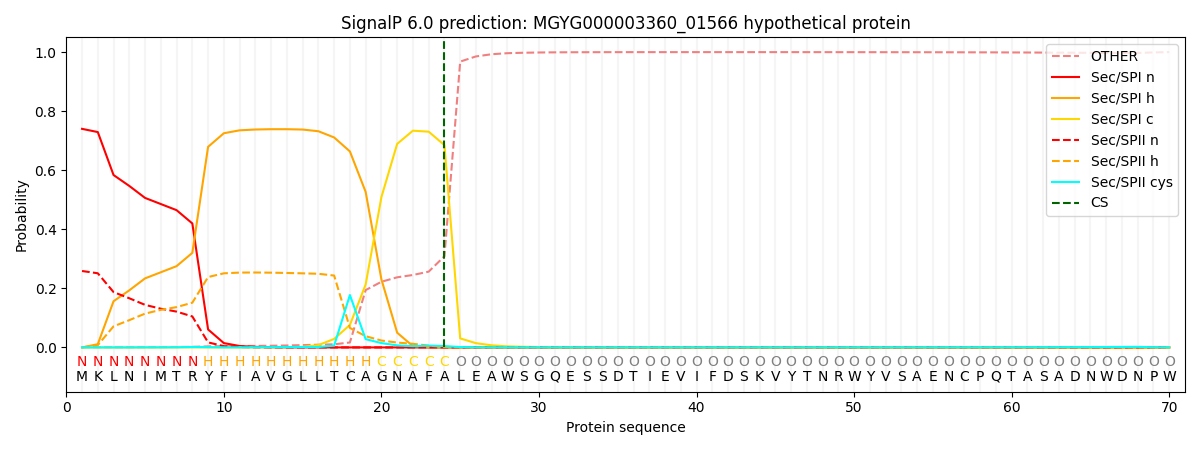
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001890 | 0.729715 | 0.267245 | 0.000587 | 0.000285 | 0.000244 |
