You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003355_00986
You are here: Home > Sequence: MGYG000003355_00986
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium phytofermentans_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; Lachnoclostridium phytofermentans_A | |||||||||||
| CAZyme ID | MGYG000003355_00986 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12162; End: 14369 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 70 | 353 | 2.4e-77 | 0.9891304347826086 |
| CBM46 | 388 | 470 | 1.3e-22 | 0.9540229885057471 |
| CBM2 | 638 | 732 | 5.3e-21 | 0.9306930693069307 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.15e-42 | 70 | 354 | 16 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.64e-30 | 28 | 364 | 33 | 371 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam00553 | CBM_2 | 1.84e-13 | 640 | 731 | 5 | 96 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| pfam18448 | CBM46 | 2.61e-13 | 476 | 581 | 2 | 105 | Carbohydrate binding domain. Carbohydrate active enzymes (CAZYmes) that target recalcitrant polysaccharides are modular enzymes containing noncatalytic carbohydrate-binding modules (CBMs) that direct enzymes to their cognate substrate, thus potentiating catalysis. The structure of Bacillus halodurans endo-beta-1,4-glucanase B (Cel5B) reveals that CBM46 is tightly associated with the catalytic module and, dependent on the glucan presented to the enzyme, can contribute directly to substrate binding or play a targeting role in directing the enzyme to regions of the plant cell wall rich in the polysaccharide hydrolyzed by the enzyme. The CBM46 domain displays a classic beta-sandwich jelly roll fold. Against beta-1,3-1,4-glucans CBM46 domain participates in productive substrate binding and thus plays a direct role in the hydrolytic activity of the enzyme. |
| smart00637 | CBD_II | 5.16e-12 | 643 | 732 | 1 | 89 | CBD_II domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX43556.1 | 0.0 | 1 | 735 | 1 | 743 |
| QTL78854.1 | 4.45e-200 | 37 | 578 | 254 | 807 |
| QTL80832.1 | 6.30e-200 | 37 | 578 | 254 | 807 |
| ADB10706.1 | 1.01e-199 | 37 | 578 | 258 | 811 |
| VEG24685.1 | 1.01e-199 | 37 | 578 | 258 | 811 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4V2X_A | 5.75e-111 | 37 | 579 | 56 | 582 | ChainA, Endo-beta-1,4-glucanase (cellulase B) [Halalkalibacterium halodurans] |
| 5E0C_A | 5.48e-107 | 40 | 578 | 11 | 533 | StructuralInsight of a Trimodular Halophilic Cellulase with a Family 46 Carbohydrate-Binding Module [Bacillus sp. BG-CS10] |
| 5XRC_A | 1.09e-106 | 15 | 578 | 21 | 566 | ATrimodular GH5_4 Subfamily Endoglucanase Structure with Large Unit Cell [Bacillus sp. BG-CS10],5XRC_B A Trimodular GH5_4 Subfamily Endoglucanase Structure with Large Unit Cell [Bacillus sp. BG-CS10],5XRC_C A Trimodular GH5_4 Subfamily Endoglucanase Structure with Large Unit Cell [Bacillus sp. BG-CS10] |
| 5E09_A | 1.46e-102 | 40 | 578 | 11 | 533 | StructuralInsight of a Trimodular Halophilic Cellulase with a Family 46 Carbohydrate-Binding Module [Bacillus sp. BG-CS10] |
| 4YZP_A | 9.18e-63 | 28 | 529 | 13 | 494 | Crystalstructure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis [Bacillus licheniformis],4YZT_A Crystal structure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis complexed with cellotetraose [Bacillus licheniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23550 | 3.96e-117 | 4 | 579 | 2 | 562 | Endoglucanase B OS=Paenibacillus lautus OX=1401 GN=celB PE=3 SV=1 |
| P54937 | 6.71e-51 | 1 | 352 | 1 | 344 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P28621 | 1.17e-49 | 37 | 353 | 40 | 341 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P28623 | 5.40e-49 | 39 | 383 | 43 | 377 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P10477 | 2.77e-45 | 49 | 353 | 67 | 352 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
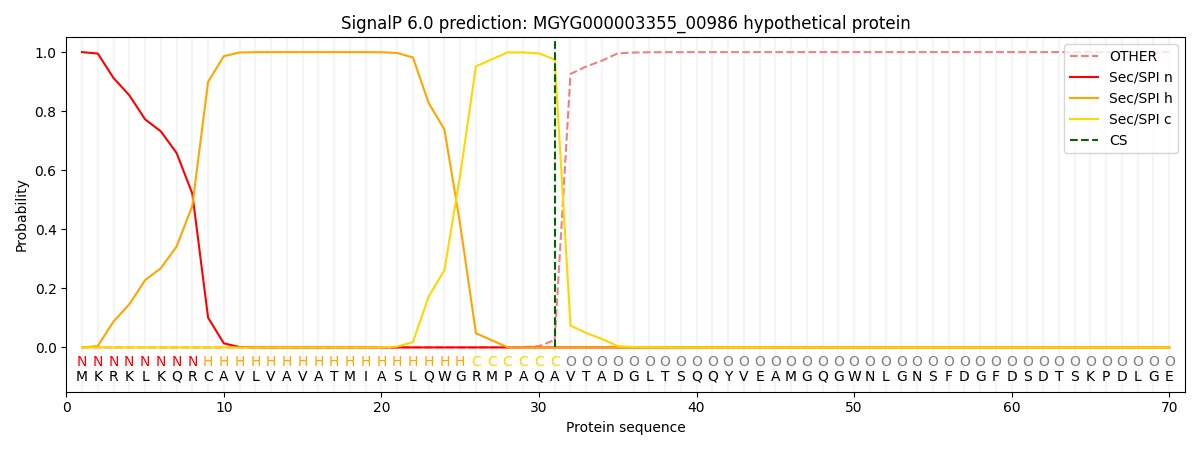
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000244 | 0.999036 | 0.000201 | 0.000190 | 0.000168 | 0.000142 |
