You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003306_01661
You are here: Home > Sequence: MGYG000003306_01661
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
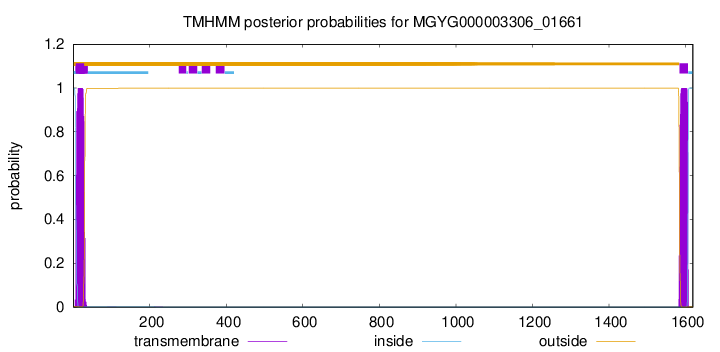
TMHMM annotations
Basic Information help
| Species | CAG-448 sp003150135 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; CAG-272; CAG-448; CAG-448 sp003150135 | |||||||||||
| CAZyme ID | MGYG000003306_01661 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 59783; End: 64642 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 64 | 728 | 6.1e-74 | 0.6795212765957447 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.26e-35 | 103 | 691 | 29 | 633 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam13385 | Laminin_G_3 | 6.48e-15 | 1136 | 1261 | 20 | 149 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| cd00063 | FN3 | 4.92e-10 | 887 | 973 | 1 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| PRK10150 | PRK10150 | 1.26e-08 | 149 | 484 | 80 | 417 | beta-D-glucuronidase; Provisional |
| pfam00041 | fn3 | 4.46e-08 | 890 | 961 | 3 | 81 | Fibronectin type III domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT26201.1 | 8.61e-170 | 18 | 881 | 11 | 855 |
| SCV07058.1 | 1.20e-169 | 18 | 881 | 11 | 855 |
| QRQ54566.1 | 1.20e-169 | 18 | 881 | 11 | 855 |
| ALJ47660.1 | 1.20e-169 | 18 | 881 | 11 | 855 |
| QRM99695.1 | 2.33e-169 | 18 | 881 | 11 | 855 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BYE_A | 4.08e-57 | 73 | 890 | 6 | 859 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
| 6BYC_A | 4.12e-57 | 73 | 890 | 6 | 859 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYI_A | 1.69e-56 | 73 | 890 | 4 | 857 | Crystalstructure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYI_B Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYG_A | 2.28e-56 | 73 | 890 | 6 | 859 | Crystalstructure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYG_B Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 5N6U_A | 3.19e-54 | 73 | 690 | 27 | 656 | Crystalstructure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5B7W2 | 6.28e-42 | 104 | 710 | 32 | 675 | Beta-mannosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=mndB PE=1 SV=2 |
| Q4FZV0 | 7.83e-42 | 74 | 694 | 24 | 695 | Beta-mannosidase OS=Rattus norvegicus OX=10116 GN=Manba PE=2 SV=1 |
| A1D911 | 5.75e-40 | 104 | 710 | 32 | 675 | Beta-mannosidase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=mndB PE=3 SV=1 |
| Q4WAH4 | 7.62e-40 | 104 | 710 | 32 | 675 | Beta-mannosidase B OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=mndB PE=3 SV=1 |
| B0YBU9 | 1.34e-39 | 104 | 710 | 32 | 675 | Beta-mannosidase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=mndB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
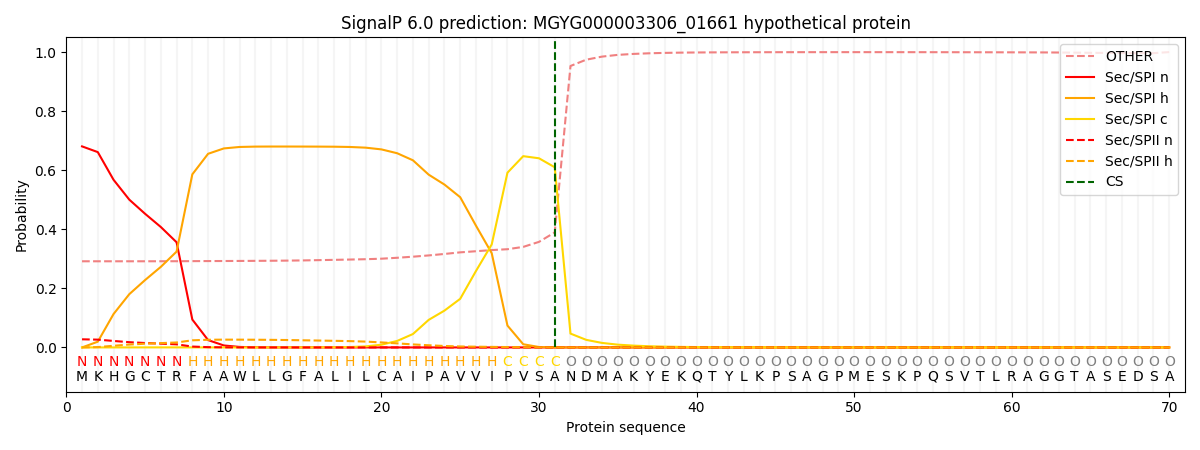
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.311698 | 0.656036 | 0.030253 | 0.000821 | 0.000472 | 0.000700 |