You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003268_01475
You are here: Home > Sequence: MGYG000003268_01475
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900762125 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900762125 | |||||||||||
| CAZyme ID | MGYG000003268_01475 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11845; End: 14121 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 46 | 277 | 1.7e-42 | 0.7743055555555556 |
| CE19 | 422 | 754 | 4e-26 | 0.7981927710843374 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02708 | PLN02708 | 8.16e-16 | 15 | 272 | 222 | 495 | Probable pectinesterase/pectinesterase inhibitor |
| pfam01095 | Pectinesterase | 1.67e-15 | 47 | 238 | 18 | 199 | Pectinesterase. |
| PLN02301 | PLN02301 | 1.27e-14 | 30 | 209 | 230 | 405 | pectinesterase/pectinesterase inhibitor |
| COG0412 | DLH | 4.32e-14 | 454 | 707 | 9 | 178 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| PLN02990 | PLN02990 | 4.49e-13 | 23 | 238 | 252 | 459 | Probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY37157.1 | 4.22e-118 | 376 | 751 | 373 | 726 |
| AGB28243.1 | 1.18e-83 | 36 | 344 | 942 | 1249 |
| VEH16322.1 | 7.92e-73 | 29 | 344 | 1016 | 1340 |
| QCD38476.1 | 2.81e-52 | 42 | 297 | 1047 | 1295 |
| QCP72166.1 | 2.81e-52 | 42 | 297 | 1047 | 1295 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XG2_A | 2.15e-13 | 33 | 209 | 7 | 172 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
| 3G8Y_A | 9.79e-07 | 404 | 745 | 45 | 352 | ChainA, SusD/RagB-associated esterase-like protein [Phocaeicola vulgatus ATCC 8482] |
| 4PMH_A | 8.58e-06 | 109 | 255 | 145 | 291 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34973 | 3.38e-29 | 411 | 753 | 2 | 299 | Putative hydrolase YtaP OS=Bacillus subtilis (strain 168) OX=224308 GN=ytaP PE=3 SV=1 |
| Q96576 | 1.83e-13 | 33 | 211 | 235 | 401 | Pectinesterase 3 OS=Solanum lycopersicum OX=4081 GN=PME3 PE=3 SV=1 |
| O04887 | 1.56e-12 | 30 | 266 | 198 | 433 | Pectinesterase 2 OS=Citrus sinensis OX=2711 GN=PECS-2.1 PE=2 SV=1 |
| P14280 | 3.93e-12 | 33 | 209 | 236 | 401 | Pectinesterase 1 OS=Solanum lycopersicum OX=4081 GN=PME1.9 PE=1 SV=5 |
| P09607 | 1.59e-11 | 56 | 209 | 257 | 405 | Pectinesterase 2.1 OS=Solanum lycopersicum OX=4081 GN=PME2.1 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
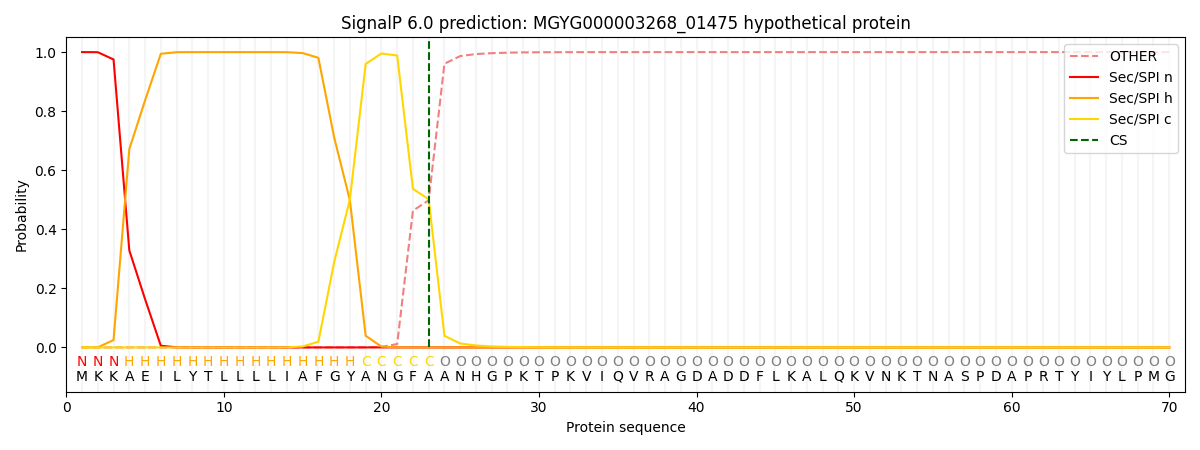
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000232 | 0.999091 | 0.000165 | 0.000173 | 0.000158 | 0.000145 |

