You are browsing environment: HUMAN GUT
MGYG000003221_00455
Basic Information
help
Species
OM05-12 sp900760755
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; OM05-12; OM05-12 sp900760755
CAZyme ID
MGYG000003221_00455
CAZy Family
GH139
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
742
85073.06
6.7328
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000003221
3509168
MAG
United States
North America
Gene Location
Start: 63799;
End: 66027
Strand: +
Family
Start
End
Evalue
family coverage
GH139
11
741
0
0.9716859716859717
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam18961
DUF5703_N
5.13e-170
26
307
1
287
Domain of unknown function (DUF5703). This is an N-terminal domain of unknown function mostly found in bacteria. It is possible that this domain might be a putative glycoside hydrolase. This family belongs to the Galactose Mutarotase-like superfamily.
more
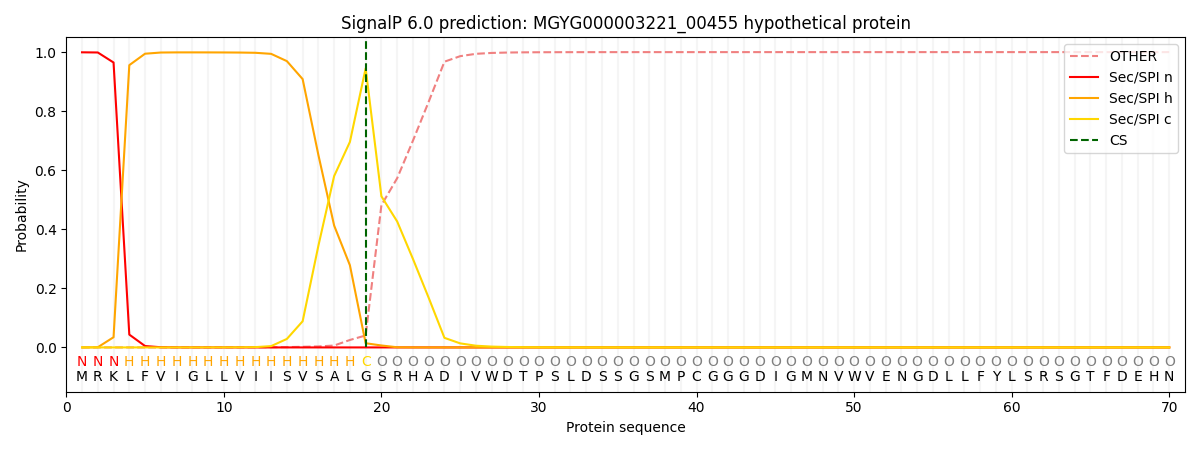
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000604
0.997562
0.001237
0.000201
0.000185
0.000181
There is no transmembrane helices in MGYG000003221_00455.