You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003172_00448
You are here: Home > Sequence: MGYG000003172_00448
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900555915 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900555915 | |||||||||||
| CAZyme ID | MGYG000003172_00448 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5765; End: 7021 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 74 | 370 | 1.5e-94 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 9.08e-53 | 63 | 371 | 4 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 7.53e-25 | 48 | 386 | 44 | 376 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT88430.1 | 3.72e-164 | 52 | 418 | 228 | 597 |
| ALJ60576.1 | 1.18e-162 | 52 | 418 | 142 | 511 |
| ADY35478.1 | 1.79e-155 | 47 | 418 | 215 | 591 |
| AVM57519.1 | 3.26e-154 | 46 | 418 | 179 | 562 |
| ADD61911.1 | 6.20e-151 | 46 | 418 | 101 | 484 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6WQY_A | 6.57e-155 | 47 | 397 | 23 | 375 | ChainA, Cellulase [Phocaeicola salanitronis DSM 18170] |
| 4YHE_A | 1.08e-134 | 48 | 418 | 7 | 385 | NativeBacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a],4YHE_B Native Bacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a] |
| 4YHG_A | 8.74e-134 | 48 | 418 | 7 | 385 | NativeBacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a],4YHG_B Native Bacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a] |
| 2JEP_A | 2.82e-87 | 44 | 404 | 31 | 394 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 6MQ4_A | 5.44e-69 | 46 | 377 | 10 | 323 | ChainA, cellulase [Acetivibrio cellulolyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O08342 | 5.52e-82 | 45 | 404 | 37 | 399 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| Q12647 | 4.19e-65 | 46 | 382 | 23 | 343 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| P28623 | 1.11e-64 | 44 | 388 | 33 | 358 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P28621 | 2.32e-62 | 46 | 388 | 40 | 360 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P23660 | 1.61e-59 | 46 | 399 | 25 | 362 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
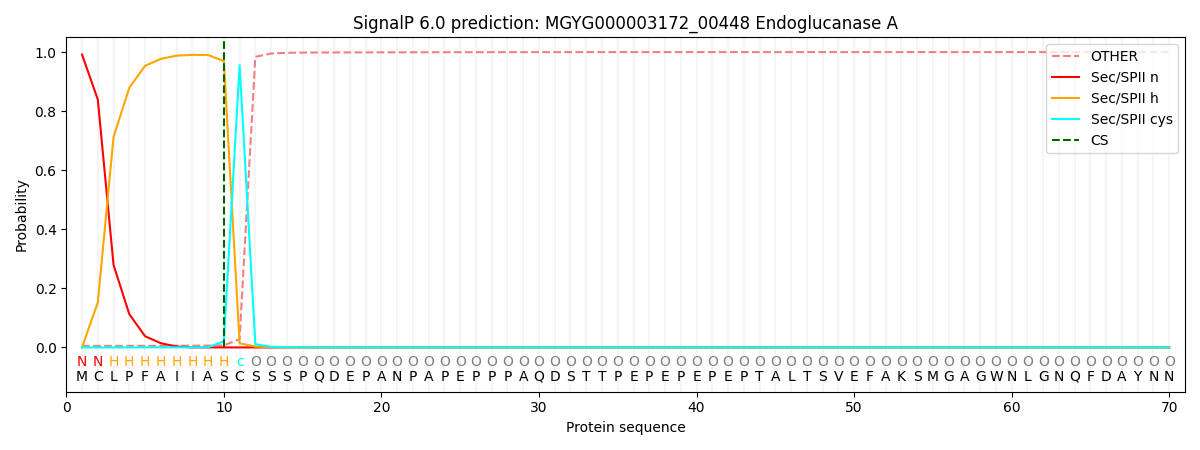
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.005317 | 0.002918 | 0.991788 | 0.000002 | 0.000004 | 0.000002 |
