You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003145_00841
You are here: Home > Sequence: MGYG000003145_00841
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS822 sp900545825 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; UMGS822; UMGS822 sp900545825 | |||||||||||
| CAZyme ID | MGYG000003145_00841 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 33544; End: 37866 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 506 | 1027 | 5.5e-82 | 0.9938900203665988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05616 | Neisseria_TspB | 3.84e-08 | 1312 | 1376 | 324 | 391 | Neisseria meningitidis TspB protein. This family consists of several Neisseria meningitidis TspB virulence factor proteins. |
| pfam05616 | Neisseria_TspB | 6.48e-08 | 1299 | 1377 | 321 | 398 | Neisseria meningitidis TspB protein. This family consists of several Neisseria meningitidis TspB virulence factor proteins. |
| pfam13229 | Beta_helix | 2.88e-07 | 700 | 864 | 9 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| NF033845 | MSCRAMM_ClfB | 3.68e-07 | 1307 | 1362 | 536 | 591 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
| NF033839 | PspC_subgroup_2 | 9.86e-07 | 1308 | 1438 | 427 | 556 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOZ72967.1 | 7.00e-171 | 448 | 1307 | 26 | 842 |
| QFU99648.1 | 2.44e-98 | 492 | 1305 | 30 | 773 |
| QGN59088.1 | 5.83e-80 | 506 | 1107 | 38 | 585 |
| QYX75179.1 | 4.15e-67 | 506 | 1220 | 37 | 689 |
| QSY58188.1 | 5.05e-55 | 360 | 1198 | 164 | 990 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6KQT_A | 5.91e-26 | 506 | 898 | 244 | 637 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 2.23e-24 | 506 | 898 | 244 | 637 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
| 7V6I_A | 1.51e-21 | 505 | 922 | 12 | 485 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 5GQC_A | 5.36e-20 | 506 | 922 | 17 | 473 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 3F2Z_A | 2.92e-06 | 270 | 410 | 16 | 136 | Crystalstructure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
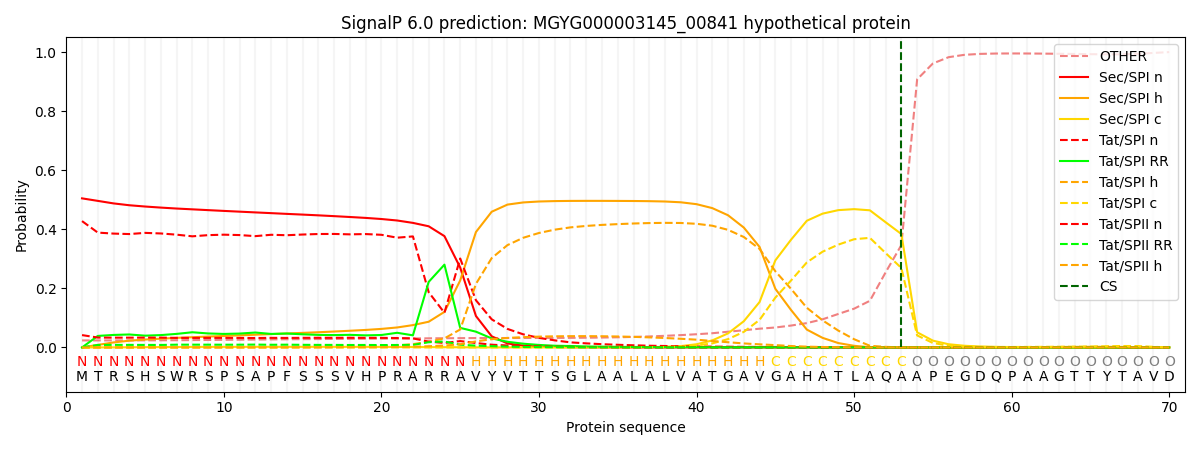
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.026964 | 0.493768 | 0.002699 | 0.433515 | 0.042680 | 0.000349 |

