You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003005_01041
You are here: Home > Sequence: MGYG000003005_01041
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
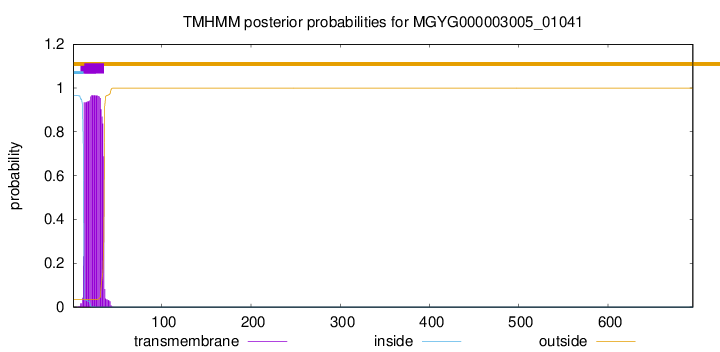
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; | |||||||||||
| CAZyme ID | MGYG000003005_01041 | |||||||||||
| CAZy Family | CBM79 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5747; End: 7834 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM79 | 431 | 553 | 4.8e-19 | 0.9636363636363636 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18522 | DUF5620 | 2.69e-21 | 247 | 407 | 1 | 119 | Domain of unknown function (DUF5620). This is a domain of unknown function predicted to be a carbohydrate binding module. |
| cd14256 | Dockerin_I | 4.87e-06 | 618 | 685 | 2 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| EWM54821.1 | 4.00e-199 | 1 | 693 | 1 | 650 |
| CBL17555.1 | 2.52e-79 | 192 | 688 | 175 | 606 |
| CCZ83178.1 | 2.35e-47 | 66 | 558 | 89 | 673 |
| CAB51934.1 | 1.78e-17 | 617 | 691 | 438 | 511 |
| AAB26620.1 | 1.78e-17 | 617 | 691 | 438 | 511 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M2S_B | 8.86e-26 | 616 | 691 | 27 | 101 | R.flavefaciens' third ScaB cohesin in complex with a group 1 dockerin [Ruminococcus flavefaciens FD-1] |
| 5M2O_B | 6.11e-12 | 618 | 688 | 29 | 90 | R.flavefaciens' third ScaB cohesin in complex with a group 1 dockerin [Ruminococcus flavefaciens FD-1] |
| 5N5P_B | 6.12e-06 | 613 | 682 | 4 | 67 | Crystalstructure of Ruminococcus flavefaciens' type III complex containing the fifth cohesin from scaffoldin B and the dockerin from scaffoldin A [Ruminococcus flavefaciens FD-1],5N5P_D Crystal structure of Ruminococcus flavefaciens' type III complex containing the fifth cohesin from scaffoldin B and the dockerin from scaffoldin A [Ruminococcus flavefaciens FD-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q53317 | 3.56e-18 | 617 | 691 | 438 | 511 | Xylanase/beta-glucanase OS=Ruminococcus flavefaciens OX=1265 GN=xynD PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
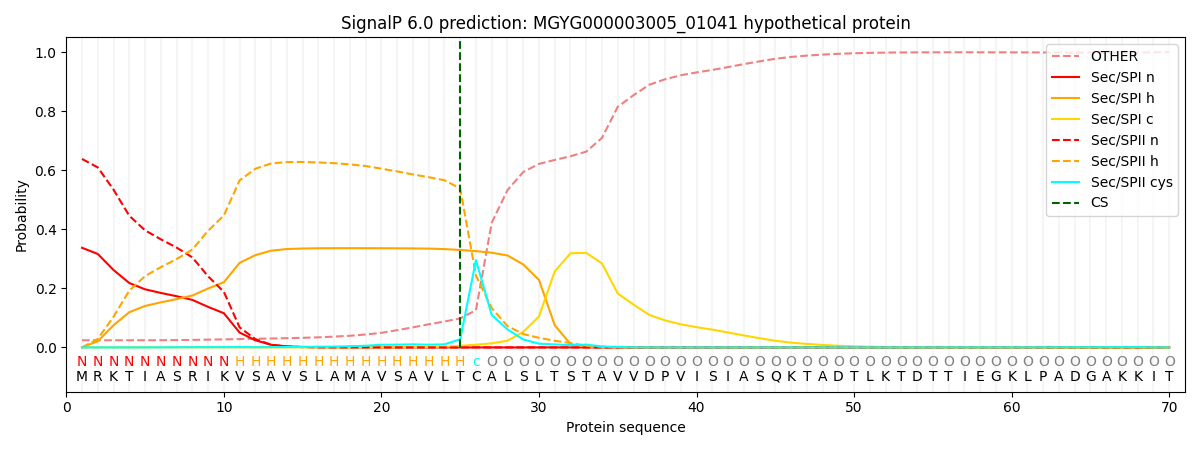
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.025715 | 0.330324 | 0.643114 | 0.000338 | 0.000226 | 0.000275 |