You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002832_01014
You are here: Home > Sequence: MGYG000002832_01014
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900548195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900548195 | |||||||||||
| CAZyme ID | MGYG000002832_01014 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9265; End: 10824 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 167 | 442 | 1.3e-70 | 0.9782608695652174 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.22e-36 | 160 | 442 | 14 | 268 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.06e-17 | 137 | 406 | 40 | 322 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGH14017.1 | 1.14e-111 | 36 | 502 | 42 | 525 |
| AHW46443.1 | 8.52e-110 | 36 | 502 | 45 | 531 |
| ADX05705.1 | 2.32e-106 | 15 | 502 | 17 | 519 |
| AIF26090.1 | 6.65e-106 | 15 | 502 | 2 | 508 |
| AHF25221.1 | 7.09e-103 | 70 | 502 | 66 | 523 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6WQY_A | 1.04e-61 | 140 | 480 | 23 | 378 | ChainA, Cellulase [Phocaeicola salanitronis DSM 18170] |
| 4YHE_A | 1.62e-61 | 139 | 502 | 5 | 386 | NativeBacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a],4YHE_B Native Bacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a] |
| 4YHG_A | 1.22e-60 | 139 | 502 | 5 | 386 | NativeBacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a],4YHG_B Native Bacteroidetes-affiliated Gh5 Cellulase Linked With A Polysaccharide Utilization Locus [Bacteroidetes bacterium AC2a] |
| 6GL2_A | 2.74e-42 | 148 | 445 | 21 | 313 | Structureof ZgEngAGH5_4 wild type at 1.2 Angstrom resolution [Zobellia galactanivorans] |
| 6UI3_A | 7.85e-42 | 140 | 454 | 15 | 317 | GH5-4broad specificity endoglucanase from Clostridum cellulovorans [Clostridium cellulovorans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q12647 | 2.79e-38 | 135 | 442 | 19 | 330 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| P20847 | 1.30e-36 | 140 | 465 | 38 | 378 | Endoglucanase 1 OS=Butyrivibrio fibrisolvens OX=831 GN=end1 PE=3 SV=1 |
| P17901 | 8.62e-36 | 108 | 499 | 12 | 418 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P28621 | 8.68e-35 | 140 | 466 | 41 | 358 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P23661 | 3.19e-33 | 140 | 442 | 70 | 367 | Endoglucanase B OS=Ruminococcus albus OX=1264 GN=celB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
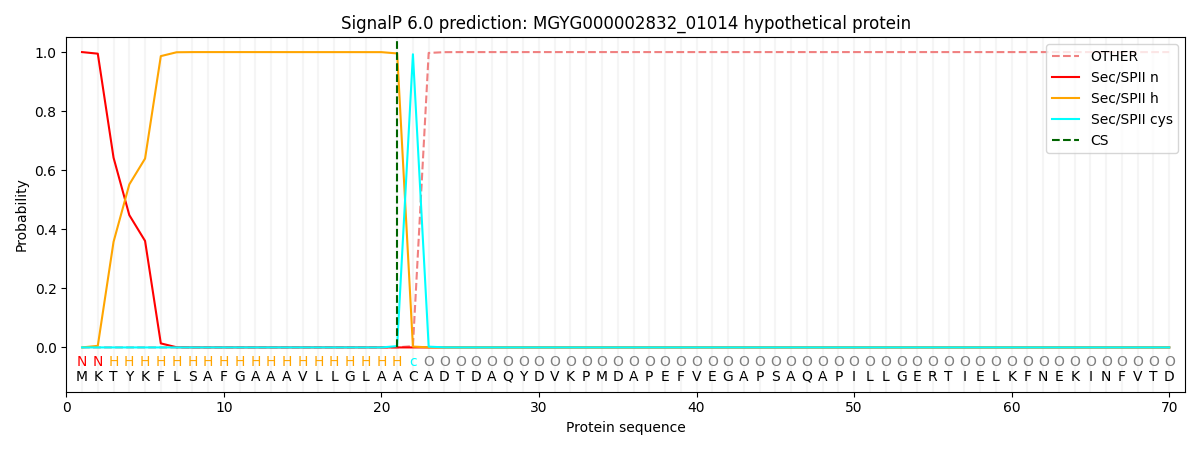
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000058 | 0.000000 | 0.000000 | 0.000000 |
