You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002832_00807
You are here: Home > Sequence: MGYG000002832_00807
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
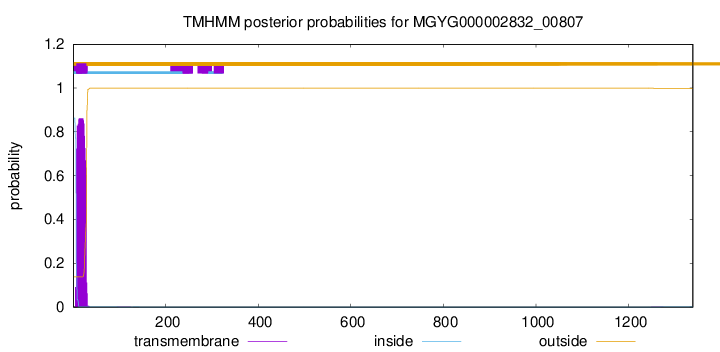
TMHMM annotations
Basic Information help
| Species | Prevotella sp900548195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900548195 | |||||||||||
| CAZyme ID | MGYG000002832_00807 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13854; End: 17873 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 81 | 258 | 6.7e-50 | 0.9890710382513661 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15319 | PRK15319 | 2.75e-13 | 593 | 1039 | 621 | 1063 | fibronectin-binding autotransporter adhesin ShdA. |
| cd00063 | FN3 | 1.16e-10 | 438 | 507 | 4 | 77 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 4.34e-09 | 438 | 507 | 4 | 77 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| PRK15319 | PRK15319 | 2.96e-08 | 596 | 1015 | 1234 | 1563 | fibronectin-binding autotransporter adhesin ShdA. |
| PRK15319 | PRK15319 | 3.94e-08 | 542 | 998 | 894 | 1326 | fibronectin-binding autotransporter adhesin ShdA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEV97716.1 | 6.08e-242 | 3 | 1090 | 2 | 1105 |
| QUT73817.1 | 4.03e-134 | 25 | 517 | 21 | 549 |
| QCD38445.1 | 5.09e-128 | 28 | 520 | 28 | 554 |
| QUT74519.1 | 8.04e-124 | 4 | 504 | 2 | 515 |
| QCP72135.1 | 4.36e-123 | 28 | 520 | 28 | 554 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5J7K_A | 3.60e-09 | 438 | 506 | 5 | 72 | Loopgrafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_B Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_C Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_D Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_E Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_F Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_G Loop grafting onto a highly stable FN3 scaffold [synthetic construct],5J7K_H Loop grafting onto a highly stable FN3 scaffold [synthetic construct] |
| 4U3H_A | 9.42e-09 | 438 | 506 | 17 | 84 | Crystalstructure of FN3con [synthetic construct] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0CLG7 | 2.77e-104 | 28 | 426 | 21 | 413 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| A1DPF0 | 6.79e-101 | 28 | 426 | 22 | 414 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
| B8NQQ7 | 9.09e-101 | 28 | 426 | 21 | 413 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q5B297 | 3.52e-100 | 28 | 426 | 21 | 410 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| Q2UB83 | 2.30e-99 | 28 | 426 | 21 | 413 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
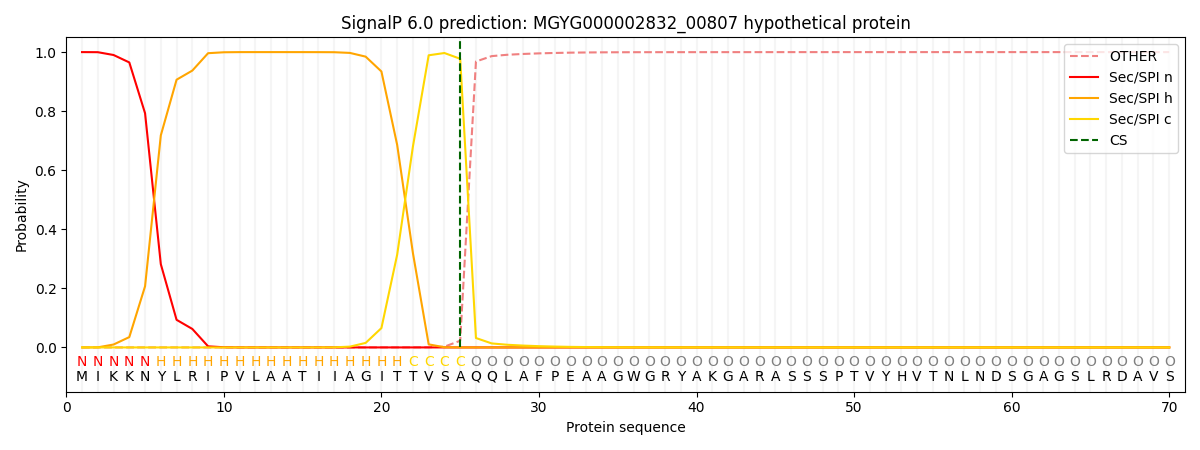
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000438 | 0.998728 | 0.000270 | 0.000183 | 0.000182 | 0.000162 |