You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002717_04036
You are here: Home > Sequence: MGYG000002717_04036
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; | |||||||||||
| CAZyme ID | MGYG000002717_04036 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 100; End: 1653 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06542 | GH18_EndoS-like | 1.31e-10 | 283 | 393 | 64 | 172 | Endo-beta-N-acetylglucosaminidases are bacterial chitinases that hydrolyze the chitin core of various asparagine (N)-linked glycans and glycoproteins. The endo-beta-N-acetylglucosaminidases have a glycosyl hydrolase family 18 (GH18) catalytic domain. Some members also have an additional C-terminal glycosyl hydrolase family 20 (GH20) domain while others have an N-terminal domain of unknown function (pfam08522). Members of this family include endo-beta-N-acetylglucosaminidase S (EndoS) from Streptococcus pyogenes, EndoF1, EndoF2, EndoF3, and EndoH from Flavobacterium meningosepticum, and EndoE from Enterococcus faecalis. EndoS is a secreted endoglycosidase from Streptococcus pyogenes that specifically hydrolyzes the glycan on human IgG between two core N-acetylglucosamine residues. EndoE is a secreted endoglycosidase, encoded by the ndoE gene in Enterococcus faecalis, that hydrolyzes the glycan on human RNase B. |
| pfam08522 | DUF1735 | 1.99e-08 | 61 | 158 | 15 | 114 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| pfam00704 | Glyco_hydro_18 | 7.56e-04 | 241 | 406 | 20 | 183 | Glycosyl hydrolases family 18. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRQ57538.1 | 0.0 | 1 | 517 | 3 | 519 |
| QDM12372.1 | 0.0 | 1 | 517 | 3 | 519 |
| SCV10295.1 | 0.0 | 1 | 517 | 3 | 519 |
| QUT81099.1 | 0.0 | 1 | 517 | 1 | 517 |
| ALJ45944.1 | 0.0 | 13 | 517 | 1 | 505 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1C8Y_A | 4.24e-12 | 244 | 416 | 33 | 206 | ChainA, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H [Streptomyces plicatus] |
| 1C3F_A | 5.71e-12 | 244 | 416 | 33 | 206 | ChainA, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H [Streptomyces plicatus] |
| 1C8X_A | 5.71e-12 | 244 | 416 | 33 | 206 | ChainA, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H [Streptomyces plicatus] |
| 1EDT_A | 8.26e-12 | 244 | 416 | 38 | 211 | ChainA, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, ENDO H [Streptomyces plicatus] |
| 6VE1_A | 9.25e-12 | 244 | 416 | 42 | 215 | ChainA, Endo-beta-N-acetylglucosaminidase H [Streptomyces plicatus],6VE1_B Chain B, Endo-beta-N-acetylglucosaminidase H [Streptomyces plicatus],6VE1_C Chain C, Endo-beta-N-acetylglucosaminidase H [Streptomyces plicatus],6VE1_D Chain D, Endo-beta-N-acetylglucosaminidase H [Streptomyces plicatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P80036 | 1.54e-12 | 244 | 427 | 83 | 261 | Endo-beta-N-acetylglucosaminidase OS=Flavobacterium sp. (strain SK1022) OX=148444 PE=1 SV=2 |
| P04067 | 6.91e-11 | 244 | 416 | 80 | 253 | Endo-beta-N-acetylglucosaminidase H OS=Streptomyces plicatus OX=1922 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
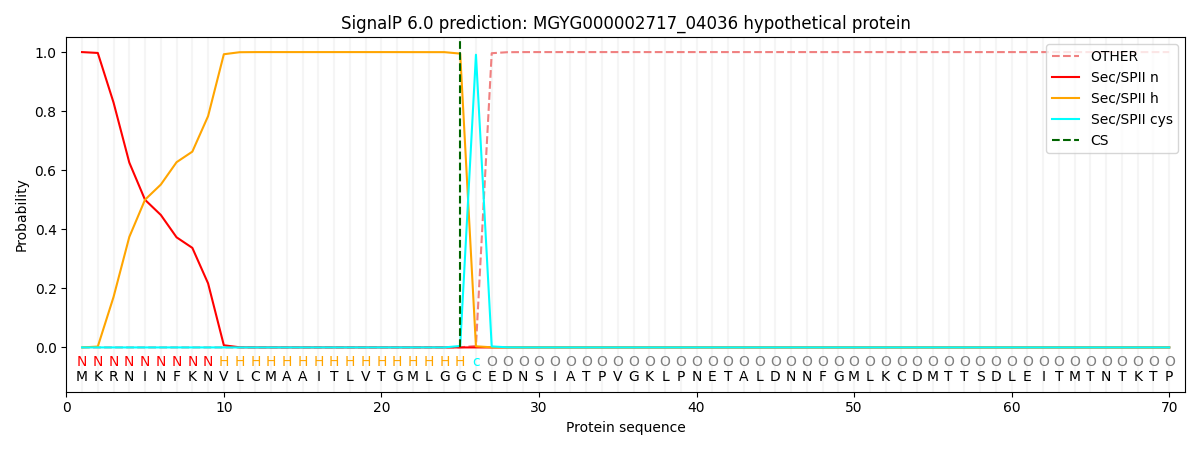
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000004 | 1.000060 | 0.000000 | 0.000000 | 0.000000 |
