You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002633_01052
You are here: Home > Sequence: MGYG000002633_01052
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
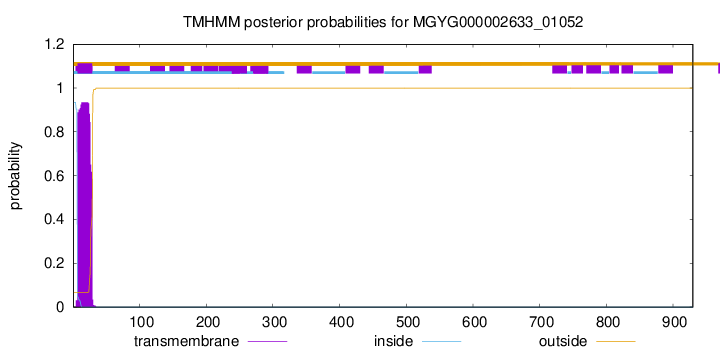
TMHMM annotations
Basic Information help
| Species | TF01-11 sp003149875 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; TF01-11; TF01-11 sp003149875 | |||||||||||
| CAZyme ID | MGYG000002633_01052 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 187; End: 2979 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 87 | 373 | 2.1e-89 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.04e-56 | 85 | 371 | 13 | 267 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 5.94e-32 | 49 | 371 | 32 | 358 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam04886 | PT | 2.51e-07 | 794 | 832 | 1 | 35 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
| pfam02368 | Big_2 | 2.04e-04 | 501 | 559 | 18 | 73 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| NF033909 | opacity_OapA | 0.002 | 789 | 904 | 233 | 346 | opacity-associated protein OapA. This family consists of full-length homologs to OapA, opacity-associated protein A as described in Haemophilus influenzae. OapA shares a C-terminal homology domain, called the OapA domain, with the Escherichia coli protein YtfB, which is now known to bind peptidoglycan through its OapA domain and to act as a cell division protein. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO18792.1 | 1.27e-133 | 52 | 735 | 246 | 944 |
| AUO19859.1 | 4.73e-85 | 48 | 614 | 101 | 687 |
| CAA73113.1 | 1.23e-80 | 1 | 385 | 1 | 381 |
| QKS59826.1 | 1.23e-80 | 1 | 385 | 1 | 381 |
| BCN29385.1 | 1.76e-80 | 56 | 756 | 349 | 974 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JEP_A | 5.39e-80 | 63 | 386 | 37 | 377 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 4X0V_A | 4.20e-62 | 63 | 385 | 42 | 377 | Structureof a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_B Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_C Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_D Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_E Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_F Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_G Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32],4X0V_H Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 [Caldicellulosiruptor sp. F32] |
| 6WQP_A | 9.00e-62 | 47 | 385 | 2 | 338 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 5H4R_A | 1.09e-61 | 63 | 385 | 42 | 377 | thecomplex of Glycoside Hydrolase 5 Lichenase from Caldicellulosiruptor sp. F32 E188Q mutant and cellotetraose [Caldicellulosiruptor sp. F32] |
| 1EDG_A | 2.75e-58 | 60 | 388 | 19 | 360 | SingleCrystal Structure Determination Of The Catalytic Domain Of Celcca Carried Out At 15 Degree C [Ruminiclostridium cellulolyticum H10] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O08342 | 2.46e-81 | 1 | 385 | 1 | 381 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P17901 | 1.58e-56 | 60 | 388 | 44 | 385 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P23661 | 8.27e-54 | 64 | 387 | 74 | 384 | Endoglucanase B OS=Ruminococcus albus OX=1264 GN=celB PE=3 SV=1 |
| P16216 | 1.05e-53 | 64 | 387 | 72 | 382 | Endoglucanase 1 OS=Ruminococcus albus OX=1264 GN=Eg I PE=1 SV=1 |
| P54937 | 2.00e-53 | 63 | 430 | 41 | 397 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
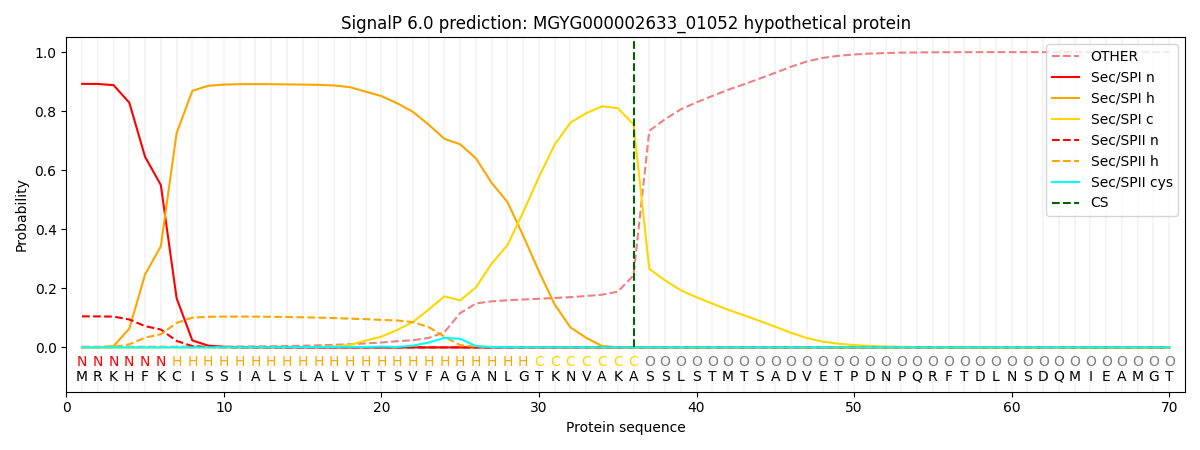
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003126 | 0.889675 | 0.106419 | 0.000343 | 0.000220 | 0.000188 |