You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002584_02136
You are here: Home > Sequence: MGYG000002584_02136
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia sp900545155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp900545155 | |||||||||||
| CAZyme ID | MGYG000002584_02136 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20839; End: 24222 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 304 | 959 | 1.3e-68 | 0.7034574468085106 |
| CBM32 | 1001 | 1121 | 3.5e-19 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.39e-30 | 304 | 714 | 9 | 414 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam00754 | F5_F8_type_C | 4.34e-19 | 998 | 1121 | 1 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| PRK10150 | PRK10150 | 8.92e-14 | 305 | 706 | 10 | 417 | beta-D-glucuronidase; Provisional |
| cd00057 | FA58C | 4.97e-10 | 992 | 1110 | 4 | 130 | Substituted updates: Jan 31, 2002 |
| pfam02836 | Glyco_hydro_2_C | 5.84e-10 | 591 | 798 | 10 | 227 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASV74706.1 | 1.79e-128 | 305 | 986 | 34 | 721 |
| QNN24101.1 | 3.45e-121 | 305 | 1125 | 40 | 841 |
| QNN24094.1 | 8.40e-115 | 302 | 986 | 49 | 725 |
| QIJ31391.1 | 4.44e-110 | 305 | 957 | 40 | 697 |
| QBN20403.1 | 2.00e-104 | 309 | 983 | 31 | 708 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D50_A | 2.27e-23 | 300 | 706 | 28 | 426 | Bacteroidesuniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii],6D50_B Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii] |
| 6D8G_A | 1.55e-22 | 300 | 706 | 28 | 426 | D341AD367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii],6D8G_B D341A D367A calcium binding mutant of Bacteroides uniformis beta-glucuronidase 2 [Bacteroides uniformis str. 3978 T3 ii] |
| 5UJ6_A | 2.65e-22 | 305 | 706 | 25 | 418 | CrystalStructure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],5UJ6_B Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],6NZG_A Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis],6NZG_B Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis] |
| 7VQM_A | 1.15e-19 | 325 | 707 | 67 | 410 | ChainA, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_B Chain B, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_C Chain C, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_D Chain D, GH2 beta-galacturonate AqGalA [Aquimarina sp.] |
| 6D8K_A | 1.04e-18 | 328 | 726 | 70 | 438 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O52847 | 2.73e-16 | 293 | 708 | 40 | 483 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q9K9C6 | 1.61e-14 | 382 | 708 | 151 | 462 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| T2KM09 | 4.82e-14 | 344 | 707 | 97 | 433 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| A7LXS9 | 2.22e-13 | 305 | 711 | 42 | 442 | Beta-galactosidase BoGH2A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02645 PE=1 SV=1 |
| B4S2K9 | 3.75e-12 | 288 | 742 | 28 | 497 | Beta-galactosidase OS=Alteromonas mediterranea (strain DSM 17117 / CIP 110805 / LMG 28347 / Deep ecotype) OX=1774373 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
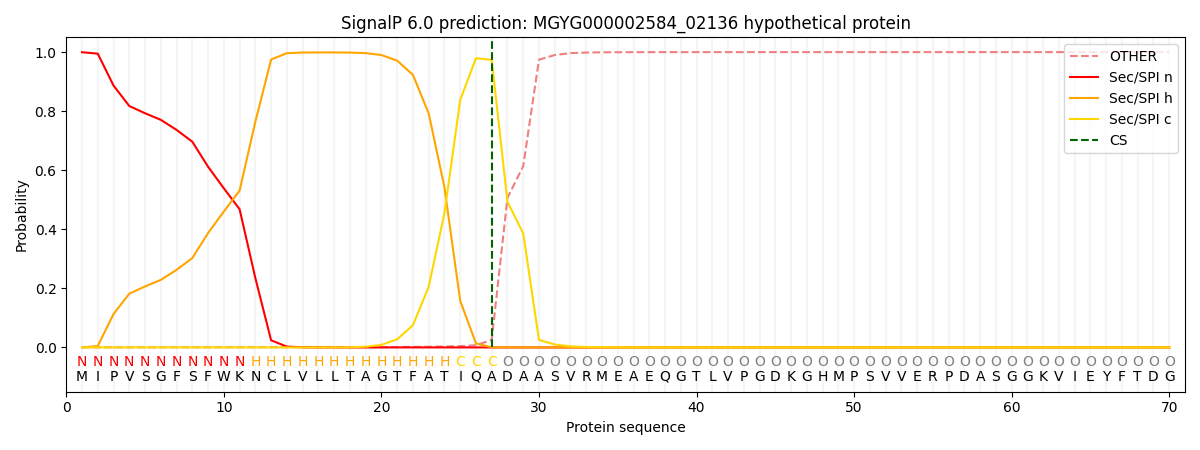
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000984 | 0.997964 | 0.000420 | 0.000235 | 0.000183 | 0.000165 |
