You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002574_01223
You are here: Home > Sequence: MGYG000002574_01223
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
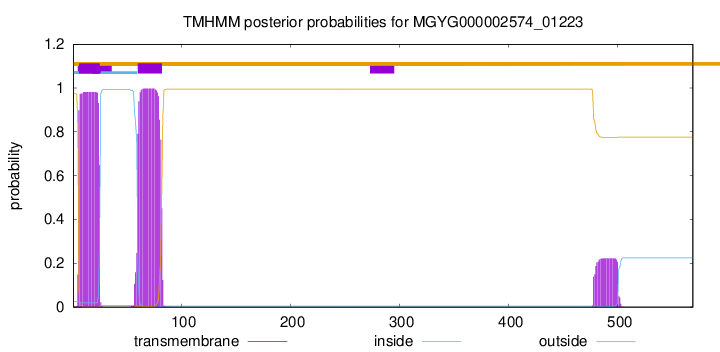
TMHMM annotations
Basic Information help
| Species | Fusobacterium_A ulcerans_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Fusobacterium_A; Fusobacterium_A ulcerans_A | |||||||||||
| CAZyme ID | MGYG000002574_01223 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1620; End: 3329 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 298 | 435 | 2e-19 | 0.7764705882352941 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04195 | GT2_AmsE_like | 8.14e-77 | 298 | 501 | 1 | 201 | GT2_AmsE_like is involved in exopolysaccharide amylovora biosynthesis. AmsE is a glycosyltransferase involved in exopolysaccharide amylovora biosynthesis in Erwinia amylovora. Amylovara is one of the three exopolysaccharide produced by E. amylovora. Amylovara-deficient mutants are non-pathogenic. It is a subfamily of Glycosyltransferase Family GT2, which includes diverse families of glycosyltransferases with a common GT-A type structural fold, which has two tightly associated beta/alpha/beta domains that tend to form a continuous central sheet of at least eight beta-strands. These are enzymes that catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. |
| pfam02397 | Bac_transf | 7.40e-71 | 55 | 234 | 1 | 180 | Bacterial sugar transferase. This Pfam family represents a conserved region from a number of different bacterial sugar transferases, involved in diverse biosynthesis pathways. |
| COG2148 | WcaJ | 9.66e-63 | 54 | 231 | 40 | 217 | Sugar transferase involved in LPS biosynthesis (colanic, teichoic acid) [Cell wall/membrane/envelope biogenesis]. |
| PRK10124 | PRK10124 | 1.36e-32 | 55 | 235 | 273 | 458 | putative UDP-glucose lipid carrier transferase; Provisional |
| PRK15204 | PRK15204 | 7.46e-30 | 52 | 231 | 277 | 467 | undecaprenyl-phosphate galactose phosphotransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARD67681.1 | 4.08e-191 | 1 | 510 | 1 | 511 |
| CCO04322.1 | 2.50e-161 | 25 | 407 | 1 | 370 |
| BAK48513.1 | 1.65e-152 | 1 | 483 | 1 | 466 |
| ALU14403.1 | 2.45e-110 | 297 | 568 | 18 | 292 |
| AXH51533.1 | 1.64e-102 | 45 | 483 | 4 | 426 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5W7L_A | 1.76e-61 | 48 | 251 | 4 | 201 | Structureof Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826],5W7L_B Structure of Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0P9D0 | 7.83e-56 | 50 | 251 | 2 | 196 | Undecaprenyl phosphate N,N'-diacetylbacillosamine 1-phosphate transferase OS=Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168) OX=192222 GN=pglC PE=1 SV=1 |
| P71062 | 1.34e-52 | 54 | 251 | 3 | 194 | Uncharacterized sugar transferase EpsL OS=Bacillus subtilis (strain 168) OX=224308 GN=epsL PE=2 SV=1 |
| Q48215 | 3.92e-46 | 297 | 566 | 3 | 267 | Uncharacterized glycosyltransferase HI_1695 OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=HI_1695 PE=3 SV=2 |
| Q46635 | 3.83e-45 | 297 | 560 | 2 | 260 | Amylovoran biosynthesis glycosyltransferase AmsE OS=Erwinia amylovora OX=552 GN=amsE PE=3 SV=2 |
| Q03084 | 1.31e-43 | 297 | 566 | 8 | 273 | UDP-Gal:alpha-D-GlcNAc-diphosphoundecaprenol beta-1,3-galactosyltransferase OS=Escherichia coli OX=562 GN=wbbD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999805 | 0.000213 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |