You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002440_02325
You are here: Home > Sequence: MGYG000002440_02325
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Enteroscipio rubneri | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Eggerthellaceae; Enteroscipio; Enteroscipio rubneri | |||||||||||
| CAZyme ID | MGYG000002440_02325 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 743; End: 3316 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 520 | 663 | 2e-26 | 0.6968085106382979 |
| CBM13 | 306 | 423 | 3.6e-24 | 0.6063829787234043 |
| CBM13 | 129 | 231 | 2e-18 | 0.526595744680851 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14200 | RicinB_lectin_2 | 4.77e-25 | 321 | 408 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 4.06e-20 | 167 | 256 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| cd16913 | YkuD_like | 3.11e-17 | 738 | 857 | 1 | 121 | L,D-transpeptidases/carboxypeptidases similar to Bacillus YkuD. Members of the YkuD-like family of proteins are found in a range of bacteria. The best studied member Bacillus YkuD has been shown to act as an L,D-transpeptidase that gives rise to an alternative pathway for peptidoglycan cross-linking. Another member Helicobacter pylori Csd6 functions as an L,D-carboxypeptidase and regulates helical cell shape and motility. The conserved region contains a conserved histidine and cysteine, with the cysteine thought to be an active site residue. |
| pfam14200 | RicinB_lectin_2 | 4.62e-17 | 609 | 696 | 2 | 88 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 5.32e-16 | 556 | 649 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRT49575.1 | 8.19e-118 | 115 | 720 | 269 | 875 |
| BAK44091.1 | 1.55e-77 | 117 | 735 | 288 | 906 |
| QRT49576.1 | 3.00e-50 | 116 | 633 | 413 | 930 |
| BAK44097.1 | 5.87e-29 | 119 | 346 | 839 | 1068 |
| AMK55176.1 | 2.72e-28 | 116 | 471 | 662 | 970 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4XXT_A | 5.60e-28 | 725 | 857 | 129 | 261 | Crystalstructure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from Clostridium acetobutylicum ATCC 824 [Clostridium acetobutylicum ATCC 824] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C0SP99 | 7.05e-08 | 671 | 856 | 7 | 193 | Putative L,D-transpeptidase YciB OS=Bacillus subtilis (strain 168) OX=224308 GN=yciB PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
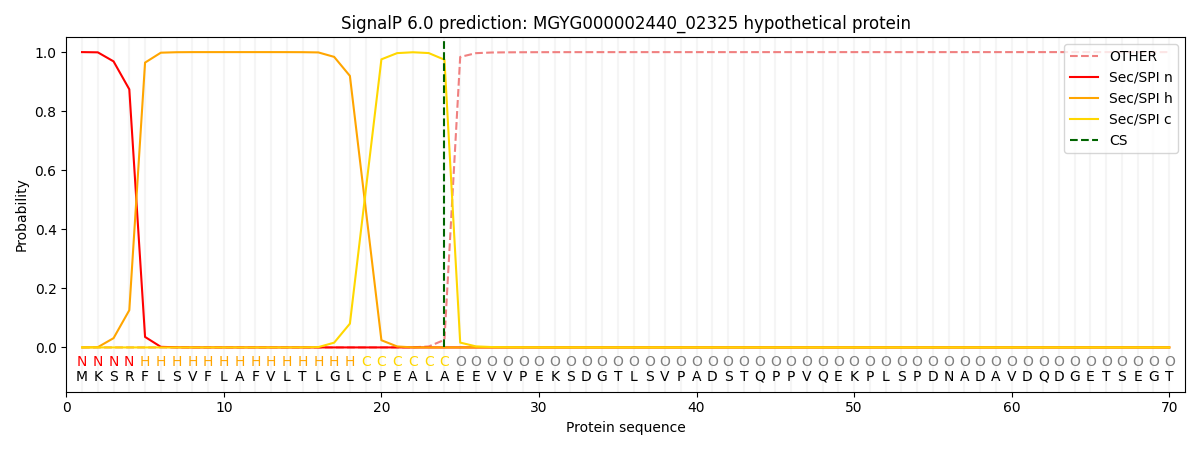
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000266 | 0.999040 | 0.000222 | 0.000160 | 0.000151 | 0.000137 |
