You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002438_04037
You are here: Home > Sequence: MGYG000002438_04037
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides distasonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides distasonis | |||||||||||
| CAZyme ID | MGYG000002438_04037 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1387704; End: 1388795 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 37 | 349 | 6.9e-90 | 0.8491620111731844 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 8.17e-51 | 51 | 315 | 35 | 304 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG4833 | COG4833 | 2.27e-27 | 51 | 320 | 46 | 306 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| COG4225 | YesR | 7.97e-06 | 225 | 309 | 32 | 100 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| pfam00759 | Glyco_hydro_9 | 1.77e-05 | 78 | 197 | 183 | 290 | Glycosyl hydrolase family 9. |
| pfam07470 | Glyco_hydro_88 | 5.30e-05 | 201 | 336 | 3 | 117 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKH96844.1 | 2.12e-279 | 1 | 363 | 1 | 363 |
| QCY55729.1 | 2.12e-279 | 1 | 363 | 1 | 363 |
| QUT53614.1 | 2.12e-279 | 1 | 363 | 1 | 363 |
| QRO16387.1 | 2.12e-279 | 1 | 363 | 1 | 363 |
| QUT18269.1 | 2.12e-279 | 1 | 363 | 1 | 363 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3K7X_A | 2.60e-36 | 104 | 321 | 87 | 299 | ChainA, Lin0763 protein [Listeria innocua] |
| 6SHD_A | 2.82e-26 | 37 | 320 | 75 | 348 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 1.01e-25 | 37 | 320 | 76 | 349 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 6SHM_A | 3.25e-24 | 37 | 320 | 76 | 349 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
| 4MU9_A | 4.46e-19 | 107 | 357 | 116 | 361 | Crystalstructure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O74556 | 6.37e-22 | 49 | 299 | 61 | 317 | Putative mannan endo-1,6-alpha-mannosidase C970.02 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPCC970.02 PE=3 SV=1 |
| Q9P6I3 | 6.21e-20 | 109 | 303 | 108 | 316 | Putative mannan endo-1,6-alpha-mannosidase C1198.07c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1198.07c PE=3 SV=2 |
| Q75DG6 | 1.33e-18 | 51 | 260 | 65 | 284 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=DCW1 PE=3 SV=2 |
| P36091 | 2.41e-18 | 51 | 297 | 65 | 318 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DCW1 PE=1 SV=1 |
| Q05031 | 3.37e-13 | 109 | 246 | 122 | 273 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DFG5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
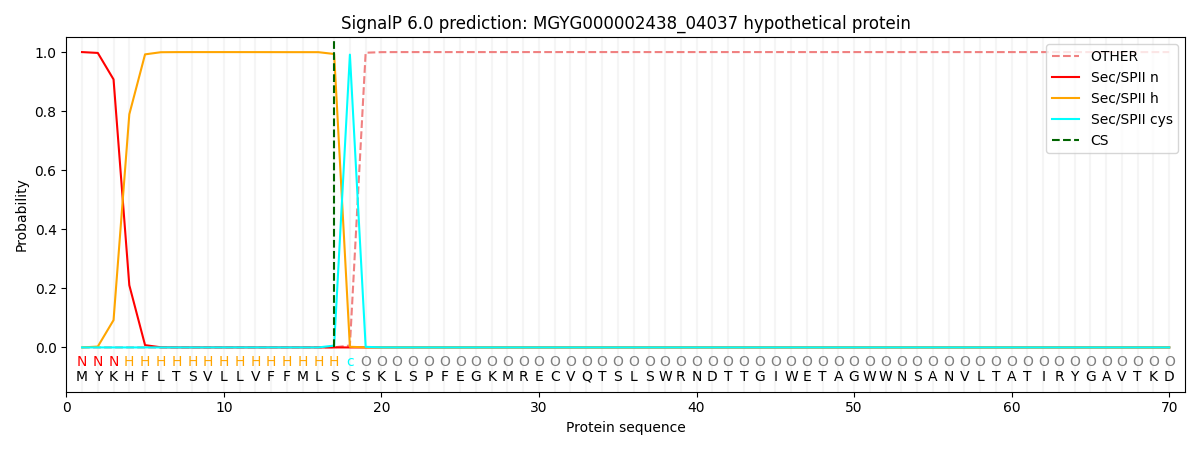
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000007 | 1.000050 | 0.000000 | 0.000000 | 0.000000 |
