You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002412_01933
You are here: Home > Sequence: MGYG000002412_01933
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotellamassilia timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotellamassilia; Prevotellamassilia timonensis | |||||||||||
| CAZyme ID | MGYG000002412_01933 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Glycosyl hydrolase family 109 protein 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 73162; End: 74826 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 61 | 474 | 2.2e-158 | 0.9899749373433584 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 2.20e-18 | 63 | 362 | 2 | 268 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 1.47e-13 | 65 | 190 | 1 | 118 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK11579 | PRK11579 | 2.14e-04 | 134 | 215 | 64 | 145 | putative oxidoreductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCQ31109.1 | 1.13e-300 | 6 | 509 | 13 | 509 |
| QFQ13386.1 | 1.02e-299 | 1 | 521 | 1 | 520 |
| QRO24012.1 | 8.10e-293 | 15 | 509 | 7 | 509 |
| ALJ59362.1 | 1.36e-282 | 1 | 501 | 1 | 498 |
| QUT89581.1 | 1.36e-282 | 1 | 501 | 1 | 498 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6T2B_A | 1.36e-83 | 47 | 473 | 25 | 438 | Glycosidehydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_B Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_C Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila],6T2B_D Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. [Akkermansia muciniphila] |
| 2IXA_A | 2.86e-73 | 65 | 477 | 21 | 435 | A-zyme,N-acetylgalactosaminidase [Elizabethkingia meningoseptica],2IXB_A Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC [Elizabethkingia meningoseptica] |
| 3E9M_A | 1.20e-06 | 101 | 239 | 39 | 173 | CrystalStructure of an oxidoreductase from Enterococcus faecalis [Enterococcus faecalis],3E9M_B Crystal Structure of an oxidoreductase from Enterococcus faecalis [Enterococcus faecalis],3E9M_C Crystal Structure of an oxidoreductase from Enterococcus faecalis [Enterococcus faecalis],3E9M_D Crystal Structure of an oxidoreductase from Enterococcus faecalis [Enterococcus faecalis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A6KWM1 | 1.67e-279 | 31 | 509 | 33 | 508 | Glycosyl hydrolase family 109 protein 4 OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=BVU_0105 PE=3 SV=1 |
| A6L1Z2 | 5.54e-279 | 23 | 509 | 26 | 509 | Glycosyl hydrolase family 109 protein 5 OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=BVU_2041 PE=3 SV=1 |
| A6KYY1 | 7.49e-265 | 14 | 531 | 11 | 525 | Glycosyl hydrolase family 109 protein 3 OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=BVU_0950 PE=3 SV=1 |
| A6KX96 | 2.06e-214 | 21 | 484 | 17 | 470 | Glycosyl hydrolase family 109 protein 1 OS=Phocaeicola vulgatus (strain ATCC 8482 / DSM 1447 / JCM 5826 / CCUG 4940 / NBRC 14291 / NCTC 11154) OX=435590 GN=BVU_0340 PE=3 SV=1 |
| Q89ZX8 | 4.78e-212 | 40 | 484 | 32 | 466 | Glycosyl hydrolase family 109 protein 1 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4243 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
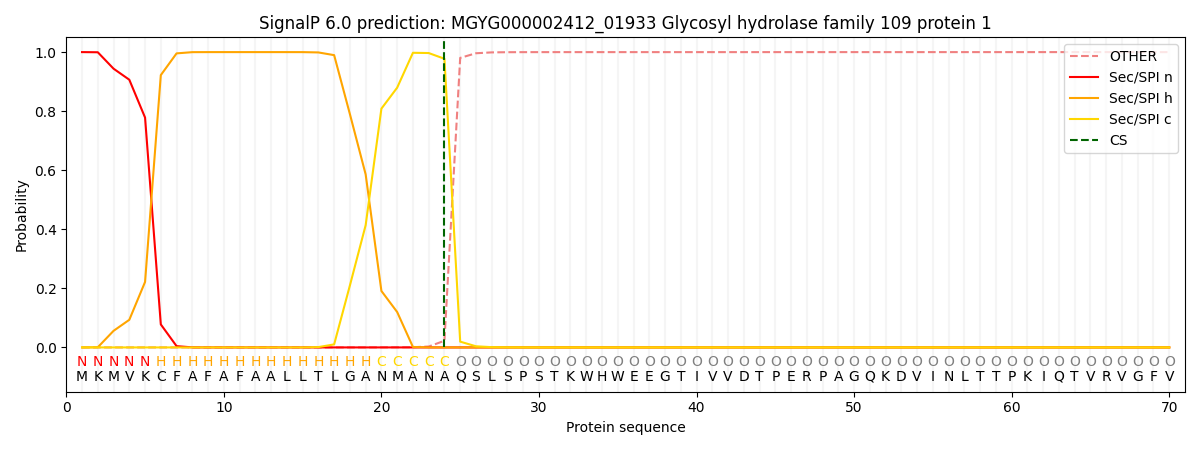
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000245 | 0.999047 | 0.000188 | 0.000183 | 0.000163 | 0.000145 |
