You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002378_02215
You are here: Home > Sequence: MGYG000002378_02215
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
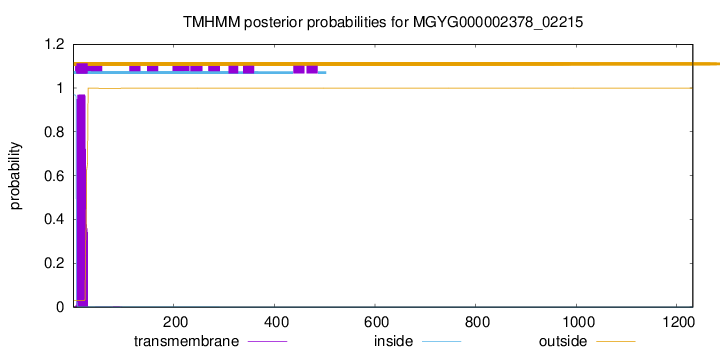
TMHMM annotations
Basic Information help
| Species | Akkermansia sp001580195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp001580195 | |||||||||||
| CAZyme ID | MGYG000002378_02215 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 248271; End: 251969 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 771 | 988 | 4e-34 | 0.7466216216216216 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00704 | Glyco_hydro_18 | 3.31e-28 | 775 | 978 | 76 | 307 | Glycosyl hydrolases family 18. |
| smart00636 | Glyco_18 | 1.00e-27 | 738 | 978 | 38 | 334 | Glyco_18 domain. |
| cd06548 | GH18_chitinase | 1.62e-16 | 769 | 978 | 85 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| pfam13385 | Laminin_G_3 | 4.44e-15 | 299 | 430 | 6 | 145 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| cd14948 | BACON | 8.04e-15 | 1014 | 1099 | 2 | 83 | Bacteroidetes-Associated Carbohydrate-binding (putative) Often N-terminal (BACON) domain. The BACON domain is found in diverse domain architectures and accociated with a wide variety of domains, including carbohydrate-active enzymes and proteases. It was named for its suggested function of carbohydrate binding; the latter was inferred from domain architectures, sequence conservation, and phyletic distribution. However, recent experimental data suggest that its primary function in Bacteroides ovatus endo-xyloglucanase BoGH5A is to distance the catalytic module from the cell surface and confer additional mobility to the catalytic domain for attack of the polysaccharide. No evidence for a direct role in carbohydrate binding could be found in that case. The large majority of BACON domains are found in Bacteroidetes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWO91303.1 | 0.0 | 3 | 1232 | 1 | 1230 |
| QWO86081.1 | 0.0 | 3 | 1232 | 1 | 1230 |
| QWO95916.1 | 0.0 | 3 | 1232 | 1 | 1230 |
| QWP70724.1 | 0.0 | 3 | 1232 | 1 | 1230 |
| QWO93387.1 | 0.0 | 3 | 1232 | 1 | 1230 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6KXN_A | 2.50e-08 | 752 | 978 | 61 | 357 | Crystalstructure of W50A mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXN_B Crystal structure of W50A mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
| 6KST_A | 2.50e-08 | 752 | 978 | 61 | 357 | Crystalstructure of the catalytic domain of chitinase ChiL from Chitiniphilus shinanonensis (CsChiL) [Chitiniphilus shinanonensis],6KST_B Crystal structure of the catalytic domain of chitinase ChiL from Chitiniphilus shinanonensis (CsChiL) [Chitiniphilus shinanonensis],6KXL_A Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXL_B Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
| 1ITX_A | 2.97e-08 | 721 | 978 | 82 | 406 | ChainA, Glycosyl Hydrolase [Niallia circulans] |
| 6KXM_A | 1.02e-07 | 752 | 978 | 61 | 357 | Crystalstructure of D157N mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXM_B Crystal structure of D157N mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
| 1D2K_A | 2.55e-07 | 775 | 978 | 99 | 348 | ChainA, CHITINASE 1 [Coccidioides immitis],1LL4_A Chain A, Chitinase 1 [Coccidioides immitis],1LL4_B Chain B, Chitinase 1 [Coccidioides immitis],1LL4_C Chain C, Chitinase 1 [Coccidioides immitis],1LL4_D Chain D, Chitinase 1 [Coccidioides immitis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 2.46e-07 | 721 | 978 | 114 | 438 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| C5P230 | 8.84e-07 | 744 | 978 | 102 | 383 | Endochitinase 1 OS=Coccidioides posadasii (strain C735) OX=222929 GN=CTS1 PE=3 SV=1 |
| P0CB51 | 1.54e-06 | 775 | 978 | 134 | 383 | Endochitinase 1 OS=Coccidioides posadasii (strain RMSCC 757 / Silveira) OX=443226 GN=CTS1 PE=1 SV=1 |
| Q1E3R8 | 1.54e-06 | 775 | 978 | 134 | 383 | Endochitinase 1 OS=Coccidioides immitis (strain RS) OX=246410 GN=CTS1 PE=3 SV=1 |
| G5EAZ3 | 2.48e-06 | 695 | 1010 | 14 | 386 | Endochitinase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=chiB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
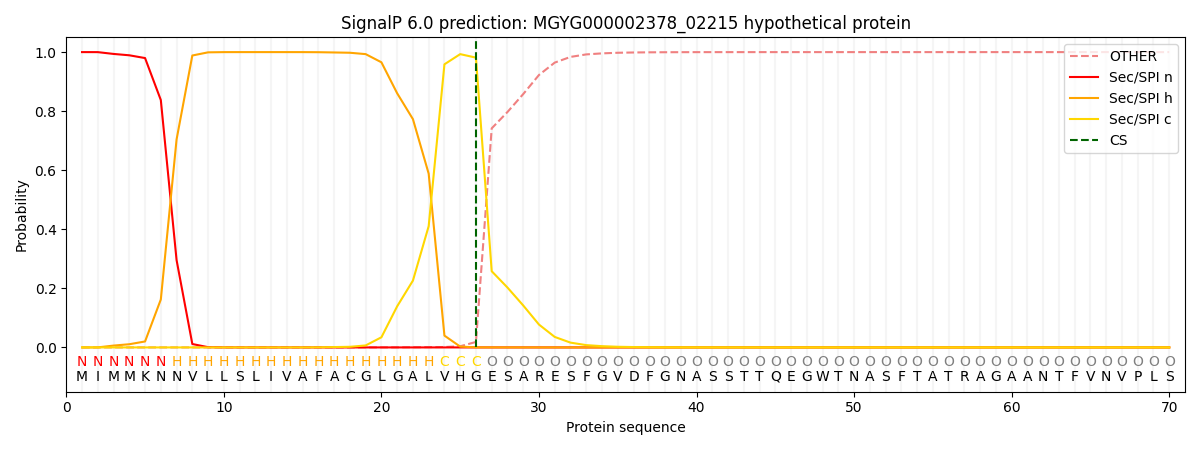
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000254 | 0.999067 | 0.000207 | 0.000157 | 0.000153 | 0.000136 |