You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002378_01068
You are here: Home > Sequence: MGYG000002378_01068
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Akkermansia sp001580195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp001580195 | |||||||||||
| CAZyme ID | MGYG000002378_01068 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1000; End: 2397 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18565 | Glyco_hydro2_C5 | 8.61e-14 | 295 | 384 | 1 | 95 | Glycoside hydrolase family 2 C-terminal domain 5. Domain 5 is found in dimeric beta-D-galactosidase from Paracoccus sp. 32d, which contributes to stabilization of the functional dimer. It is suggested that the location of this domain 5, may be one of the factors responsible for the creation of a functional dimer and cold-adaptation of this enzyme. |
| pfam02369 | Big_1 | 8.94e-05 | 309 | 381 | 1 | 64 | Bacterial Ig-like domain (group 1). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial surface proteins such as intimins and invasins involved in pathogenicity. |
| PRK13613 | PRK13613 | 4.44e-04 | 296 | 394 | 451 | 547 | lipoprotein LpqB; Provisional |
| pfam16355 | DUF4982 | 8.48e-04 | 209 | 284 | 5 | 62 | Domain of unknown function (DUF4982). This family is found in the C-terminal of uncharacterized proteins and beta-galactosidases around 680 residues in length from various Bacteroides species. The function of this protein is unknown. |
| smart00634 | BID_1 | 0.001 | 309 | 383 | 14 | 82 | Bacterial Ig-like domain (group 1). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWO94006.1 | 0.0 | 1 | 465 | 447 | 911 |
| QWO86734.1 | 0.0 | 1 | 465 | 447 | 911 |
| QWP71339.1 | 0.0 | 1 | 465 | 447 | 911 |
| QWO90637.1 | 0.0 | 1 | 465 | 447 | 911 |
| QWO96572.1 | 0.0 | 1 | 465 | 447 | 911 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EUV_A | 8.76e-10 | 1 | 395 | 346 | 728 | ChainA, Beta-D-galactosidase [Paracoccus sp. 32d],5EUV_B Chain B, Beta-D-galactosidase [Paracoccus sp. 32d],5LDR_B Chain B, Beta-D-galactosidase [Paracoccus sp. 32d] |
| 5LDR_A | 8.77e-10 | 1 | 395 | 347 | 729 | ChainA, Beta-D-galactosidase [Paracoccus sp. 32d] |
| 4YPJ_A | 6.43e-09 | 142 | 383 | 552 | 795 | ChainA, Beta galactosidase [Niallia circulans],4YPJ_B Chain B, Beta galactosidase [Niallia circulans] |
| 7CWD_A | 2.38e-07 | 140 | 384 | 543 | 790 | ChainA, beta-glalactosidase [Niallia circulans],7CWI_A Chain A, beta-galactosidase [Niallia circulans] |
| 6D7J_A | 8.77e-06 | 163 | 387 | 613 | 818 | TheCrystal Structure of Parabacteroides merdae Beta-Glucuronidase (GUS) with Glycerol in Active-Site [Parabacteroides merdae CL03T12C32],6D7J_B The Crystal Structure of Parabacteroides merdae Beta-Glucuronidase (GUS) with Glycerol in Active-Site [Parabacteroides merdae CL03T12C32],6D7J_C The Crystal Structure of Parabacteroides merdae Beta-Glucuronidase (GUS) with Glycerol in Active-Site [Parabacteroides merdae CL03T12C32],6D7J_D The Crystal Structure of Parabacteroides merdae Beta-Glucuronidase (GUS) with Glycerol in Active-Site [Parabacteroides merdae CL03T12C32],6DXU_A Crystal Structure of Parabacteroides merdae Beta-Glucuronidase (GUS) [Parabacteroides merdae ATCC 43184],6DXU_B Crystal Structure of Parabacteroides merdae Beta-Glucuronidase (GUS) [Parabacteroides merdae ATCC 43184],6DXU_C Crystal Structure of Parabacteroides merdae Beta-Glucuronidase (GUS) [Parabacteroides merdae ATCC 43184],6DXU_D Crystal Structure of Parabacteroides merdae Beta-Glucuronidase (GUS) [Parabacteroides merdae ATCC 43184] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P77989 | 2.30e-18 | 1 | 393 | 369 | 735 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| P26257 | 1.45e-08 | 1 | 346 | 370 | 704 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
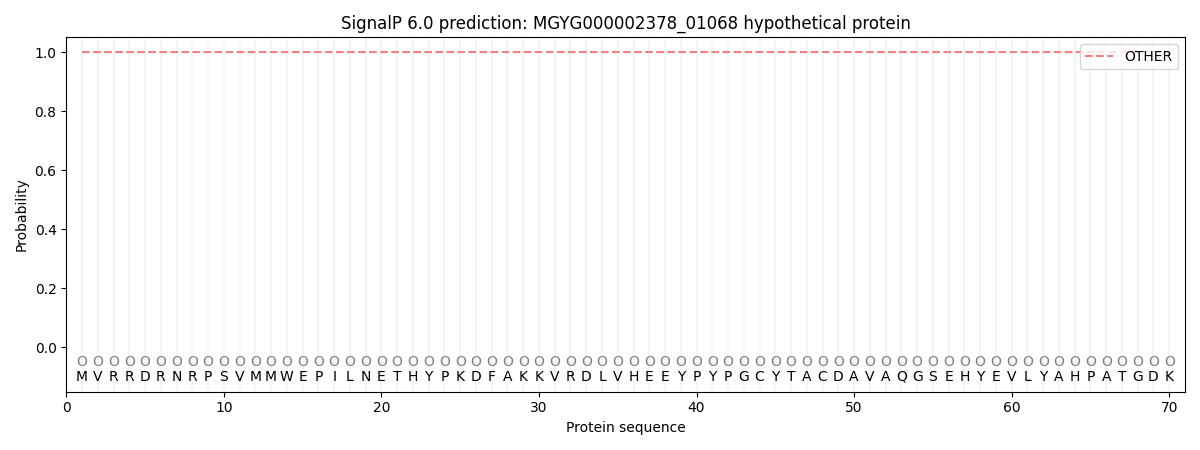
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000062 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
