You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002376_02421
You are here: Home > Sequence: MGYG000002376_02421
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
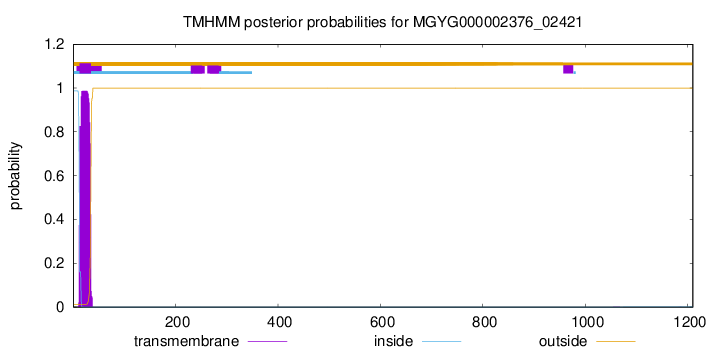
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium_B sp000765215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium_B; Lachnoclostridium_B sp000765215 | |||||||||||
| CAZyme ID | MGYG000002376_02421 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12586; End: 16218 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 150 | 308 | 1.1e-24 | 0.7978723404255319 |
| CBM13 | 846 | 992 | 1.6e-22 | 0.7180851063829787 |
| CBM13 | 456 | 546 | 1.1e-19 | 0.4627659574468085 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14200 | RicinB_lectin_2 | 7.84e-23 | 494 | 582 | 2 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 3.15e-19 | 185 | 270 | 2 | 86 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 3.32e-18 | 457 | 534 | 13 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 2.03e-17 | 881 | 972 | 2 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 4.73e-14 | 653 | 720 | 7 | 74 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRT49576.1 | 0.0 | 130 | 1210 | 106 | 1184 |
| QRT49575.1 | 3.66e-66 | 440 | 947 | 270 | 779 |
| BAK44091.1 | 7.64e-46 | 454 | 956 | 299 | 804 |
| AMK55176.1 | 8.12e-26 | 454 | 890 | 512 | 975 |
| QRT50581.1 | 7.96e-24 | 416 | 947 | 100 | 779 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
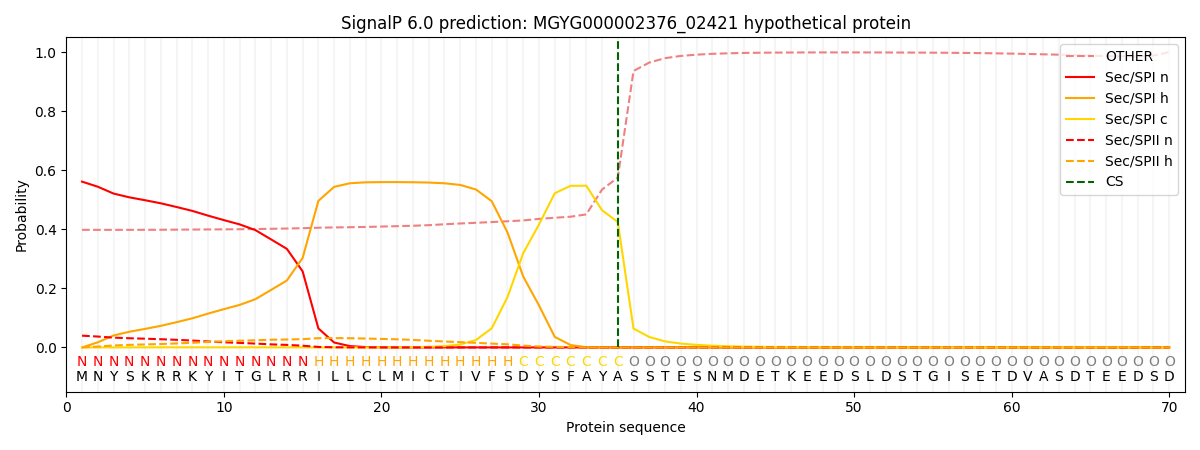
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.425032 | 0.522295 | 0.043913 | 0.003541 | 0.001884 | 0.003336 |