You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002365_00525
You are here: Home > Sequence: MGYG000002365_00525
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bifidobacterium globosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium globosum | |||||||||||
| CAZyme ID | MGYG000002365_00525 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 602506; End: 603399 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 97 | 292 | 5.4e-39 | 0.9735449735449735 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01083 | Cutinase | 1.91e-18 | 100 | 280 | 7 | 161 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASW23299.1 | 5.44e-201 | 1 | 297 | 1 | 297 |
| ATO40344.1 | 5.09e-174 | 50 | 297 | 2 | 249 |
| AIZ15958.1 | 1.56e-163 | 70 | 297 | 1 | 228 |
| AFI62706.1 | 1.49e-81 | 65 | 295 | 73 | 303 |
| AYN23342.1 | 2.24e-81 | 65 | 295 | 86 | 316 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2AXE_A | 1.59e-08 | 143 | 278 | 58 | 192 | IodinatedComplex Of Acetyl Xylan Esterase At 1.80 Angstroms [Talaromyces purpureogenus] |
| 1BS9_A | 2.15e-08 | 143 | 278 | 58 | 192 | AcetylxylanEsterase From P. Purpurogenum Refined At 1.10 Angstroms [Talaromyces purpureogenus] |
| 1G66_A | 2.15e-08 | 143 | 278 | 58 | 192 | ACETYLXYLANESTERASE AT 0.90 ANGSTROM RESOLUTION [Talaromyces purpureogenus] |
| 1QOZ_A | 9.03e-06 | 127 | 276 | 47 | 190 | ChainA, ACETYL XYLAN ESTERASE [Trichoderma reesei],1QOZ_B Chain B, ACETYL XYLAN ESTERASE [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A6WFI5 | 6.59e-12 | 100 | 295 | 41 | 220 | Cutinase OS=Kineococcus radiotolerans (strain ATCC BAA-149 / DSM 14245 / SRS30216) OX=266940 GN=cut PE=1 SV=1 |
| P63880 | 6.83e-12 | 82 | 293 | 22 | 213 | Carboxylesterase Culp1 OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=BQ2027_MB2006C PE=3 SV=1 |
| P9WP42 | 6.83e-12 | 82 | 293 | 22 | 213 | Carboxylesterase Culp1 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT2037 PE=3 SV=1 |
| P9WP43 | 6.83e-12 | 82 | 293 | 22 | 213 | Carboxylesterase Culp1 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut7 PE=1 SV=1 |
| O06793 | 1.16e-11 | 122 | 293 | 13 | 173 | Probable carboxylesterase Culp5 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
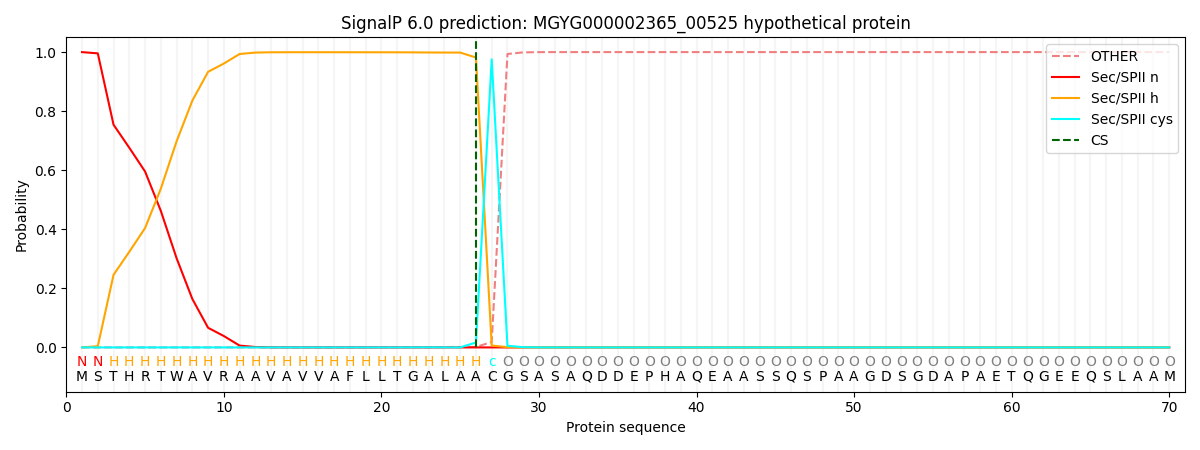
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000102 | 0.999926 | 0.000000 | 0.000000 | 0.000000 |

