You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002359_01714
Basic Information
help
| Species |
Mycobacterium tuberculosis
|
| Lineage |
Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Mycobacterium; Mycobacterium tuberculosis
|
| CAZyme ID |
MGYG000002359_01714
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000002359 |
4387829 |
Isolate |
Sweden |
Europe |
|
| Gene Location |
Start: 197034;
End: 198602
Strand: -
|
No EC number prediction in MGYG000002359_01714.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
80 |
384 |
1e-32 |
0.5537037037037037 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG5305
|
COG5305 |
4.72e-147 |
1 |
521 |
24 |
551 |
Uncharacterized membrane protein [Function unknown]. |
| pfam13231
|
PMT_2 |
1.49e-04 |
80 |
230 |
5 |
155 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG0474
|
MgtA |
0.002 |
121 |
248 |
785 |
917 |
Magnesium-transporting ATPase (P-type) [Inorganic ion transport and metabolism]. |
| COG1807
|
ArnT |
0.002 |
80 |
375 |
67 |
373 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
This protein is predicted as OTHER
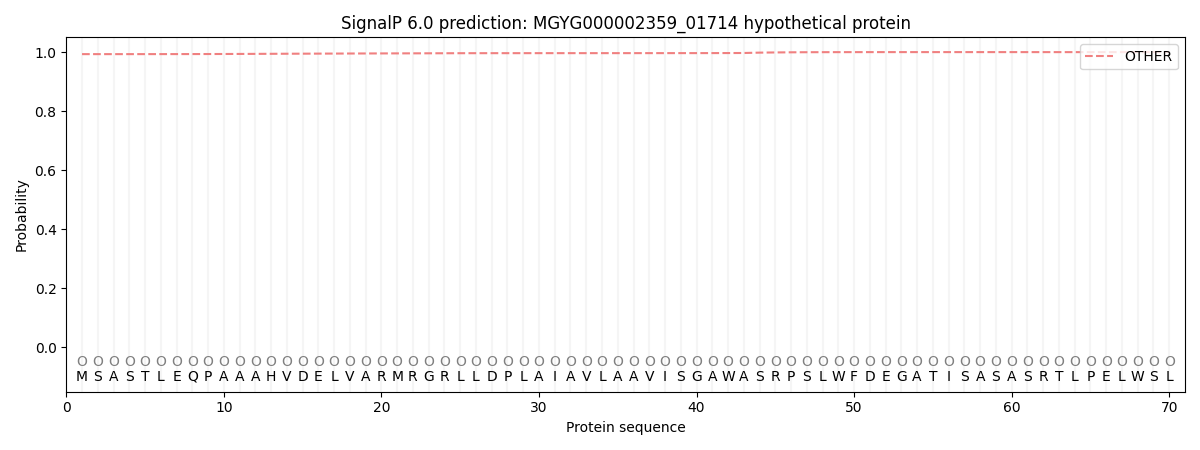
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.993825
|
0.005826
|
0.000090
|
0.000032
|
0.000019
|
0.000234
|
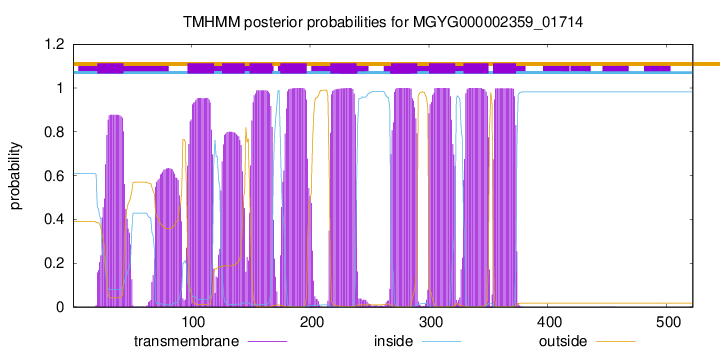
| start |
end |
| 21 |
43 |
| 97 |
119 |
| 126 |
145 |
| 149 |
168 |
| 175 |
197 |
| 217 |
239 |
| 268 |
290 |
| 300 |
322 |
| 329 |
350 |
| 354 |
373 |


