You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002348_01342
You are here: Home > Sequence: MGYG000002348_01342
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
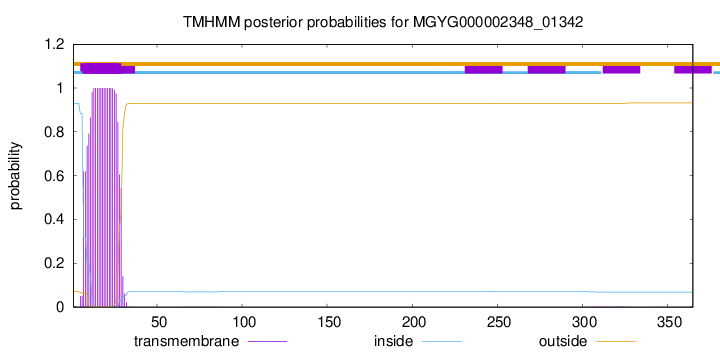
TMHMM annotations
Basic Information help
| Species | Lactobacillus crispatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lactobacillus; Lactobacillus crispatus | |||||||||||
| CAZyme ID | MGYG000002348_01342 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 37570; End: 38667 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 2.36e-19 | 40 | 361 | 8 | 315 | Glycosyl hydrolases family 8. |
| COG3405 | BcsZ | 4.91e-16 | 20 | 363 | 8 | 341 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| PRK11097 | PRK11097 | 1.92e-12 | 39 | 163 | 27 | 150 | cellulase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLL74157.1 | 5.54e-267 | 1 | 365 | 1 | 365 |
| QPP17581.1 | 6.47e-266 | 1 | 365 | 1 | 365 |
| QLK31971.1 | 7.55e-265 | 1 | 365 | 1 | 365 |
| QHQ68201.1 | 4.37e-264 | 1 | 365 | 1 | 365 |
| QGY94787.1 | 1.25e-263 | 1 | 365 | 1 | 365 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XD0_A | 1.07e-32 | 43 | 319 | 69 | 359 | ApoStructure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4],5XD0_B Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4] |
| 1V5C_A | 1.35e-22 | 37 | 323 | 32 | 339 | Thecrystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 [Bacillus sp. (in: Bacteria)],1V5D_A The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)],1V5D_B The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)] |
| 7CJU_A | 1.50e-22 | 37 | 323 | 38 | 345 | Crystalstructure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7CJU_B Crystal structure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7XGQ_A Chain A, chitosanase [Bacillus sp. K17-2],7XGQ_B Chain B, chitosanase [Bacillus sp. K17-2] |
| 6VC5_A | 1.28e-10 | 65 | 273 | 31 | 231 | 1.6Angstrom Resolution Crystal Structure of endoglucanase from Komagataeibacter sucrofermentans [Komagataeibacter sucrofermentans] |
| 1WZZ_A | 1.44e-08 | 65 | 271 | 46 | 244 | Structureof endo-beta-1,4-glucanase CMCax from Acetobacter xylinum [Komagataeibacter xylinus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19254 | 3.16e-33 | 43 | 319 | 69 | 359 | Beta-glucanase OS=Niallia circulans OX=1397 GN=bgc PE=3 SV=1 |
| P29019 | 5.31e-20 | 37 | 323 | 88 | 395 | Endoglucanase OS=Bacillus sp. (strain KSM-330) OX=72575 PE=1 SV=1 |
| P37696 | 6.02e-09 | 65 | 271 | 54 | 252 | Probable endoglucanase OS=Komagataeibacter hansenii OX=436 GN=cmcAX PE=1 SV=1 |
| Q8RSY9 | 3.09e-08 | 22 | 163 | 14 | 153 | Endoglucanase OS=Pseudomonas fluorescens (strain SBW25) OX=216595 GN=bcsZ PE=3 SV=2 |
| Q8ZLB7 | 3.77e-07 | 40 | 163 | 29 | 151 | Endoglucanase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bcsZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
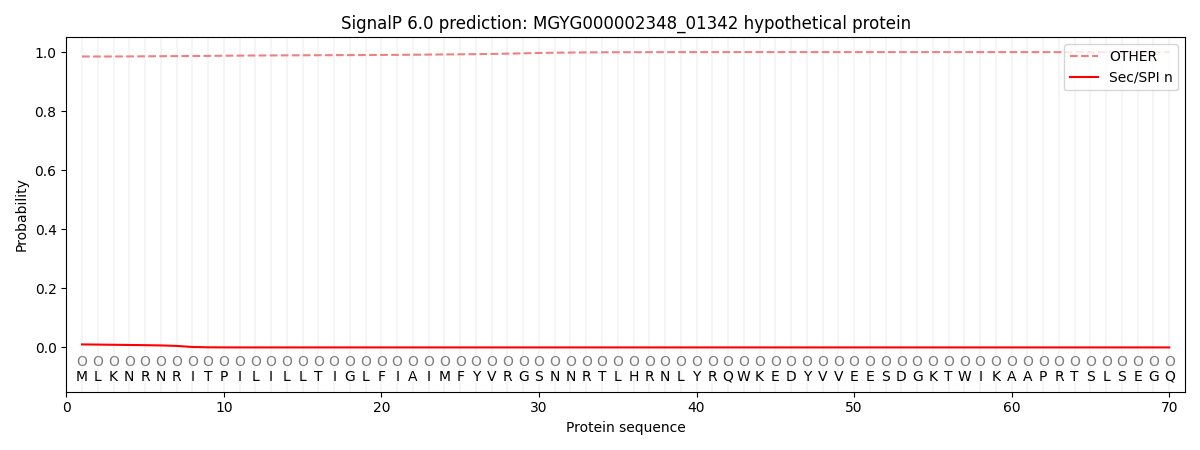
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.985667 | 0.009395 | 0.001374 | 0.000068 | 0.000035 | 0.003490 |