You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002338_01367
You are here: Home > Sequence: MGYG000002338_01367
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Staphylococcus haemolyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus haemolyticus | |||||||||||
| CAZyme ID | MGYG000002338_01367 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | putative transglycosylase SceD | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4251; End: 4958 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam06737 | Transglycosylas | 1.18e-20 | 162 | 235 | 4 | 75 | Transglycosylase-like domain. This family of proteins are very likely to act as transglycosylase enzymes related to pfam00062 and pfam01464. These other families are weakly matched by this family, and include the known active site residues. |
| cd13925 | RPF | 3.90e-18 | 166 | 234 | 6 | 70 | core lysozyme-like domain of resuscitation-promoting factor proteins. Resuscitation-promoting factor (RPF) proteins, found in various (G+C)-rich Gram-positive bacteria, act to reactivate cultures from stationary phase. This protein shares elements of the structural core of lysozyme and related proteins. Furthermore, it shares a conserved active site glutamate which is required for activity, and has a polysaccharide binding cleft that corresponds to the peptidoglycan binding cleft of lysozyme. Muralytic activity of Rpf in Micrococcus luteus correlates with resuscitation, supporting a mechanism dependent on cleavage of peptidoglycan by RPF. |
| cd00736 | lambda_lys-like | 3.55e-04 | 182 | 229 | 51 | 94 | Bacteriophage lambda lysozyme and similar proteins. Lysozyme from bacteriophage lambda hydrolyzes the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetylglucosamine (GlcNAc), as do other lysozymes. However, unlike other lysozymes, bacteriophage lambda does not produce a reducing end upon cleavage of the peptidoglycan, but rather uses the 6-OH of the same MurNAc residue to produce a 1,6-anhydromuramic acid terminal residue and is therefore a lytic transglycosylase. An identical 1,6-anhydro bond is formed in bacterial peptidoglycans by the action of the lytic transglycosylases of E. coli, though they differ structurally. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJI70515.1 | 1.04e-126 | 1 | 235 | 1 | 235 |
| AMW23878.1 | 1.04e-126 | 1 | 235 | 1 | 235 |
| AKC75696.1 | 1.04e-126 | 1 | 235 | 1 | 235 |
| BAE04248.1 | 1.04e-126 | 1 | 235 | 1 | 235 |
| QFR07441.1 | 1.04e-126 | 1 | 235 | 1 | 235 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4L7X7 | 2.08e-127 | 1 | 235 | 1 | 235 | Probable transglycosylase SceD OS=Staphylococcus haemolyticus (strain JCSC1435) OX=279808 GN=sceD PE=3 SV=1 |
| Q5HEA4 | 1.98e-84 | 1 | 235 | 1 | 231 | Probable transglycosylase SceD OS=Staphylococcus aureus (strain COL) OX=93062 GN=sceD PE=1 SV=1 |
| Q2YUK8 | 3.99e-84 | 1 | 235 | 1 | 231 | Probable transglycosylase SceD OS=Staphylococcus aureus (strain bovine RF122 / ET3-1) OX=273036 GN=sceD PE=3 SV=1 |
| Q6GEX9 | 5.66e-84 | 1 | 235 | 1 | 231 | Probable transglycosylase SceD OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=sceD PE=3 SV=1 |
| Q2FWF8 | 1.14e-83 | 1 | 235 | 1 | 231 | Probable transglycosylase SceD OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=sceD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
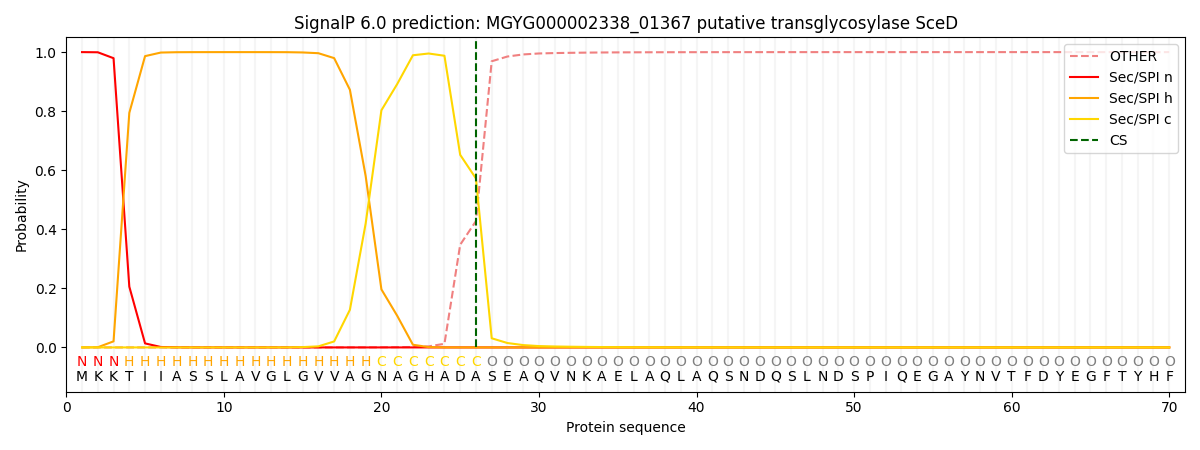
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000579 | 0.998417 | 0.000258 | 0.000265 | 0.000226 | 0.000208 |
