You are browsing environment: HUMAN GUT
MGYG000002203_01781
Basic Information
help
Species
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes;
CAZyme ID
MGYG000002203_01781
CAZy Family
GT22
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000002203
2806770
MAG
Spain
Europe
Gene Location
Start: 49577;
End: 51103
Strand: +
No EC number prediction in MGYG000002203_01781.
Family
Start
End
Evalue
family coverage
GT22
16
369
3.4e-42
0.8740359897172236
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam03901
Glyco_transf_22
8.26e-17
13
321
1
314
Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis.
more
PLN02816
PLN02816
2.50e-09
26
321
53
341
mannosyltransferase
more
COG1807
ArnT
3.04e-04
165
367
151
367
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
Q1LZA0
4.47e-08
36
269
75
302
GPI mannosyltransferase 3 OS=Bos taurus OX=9913 GN=PIGB PE=2 SV=1
more
Q92521
2.96e-06
36
277
85
321
GPI mannosyltransferase 3 OS=Homo sapiens OX=9606 GN=PIGB PE=1 SV=1
more
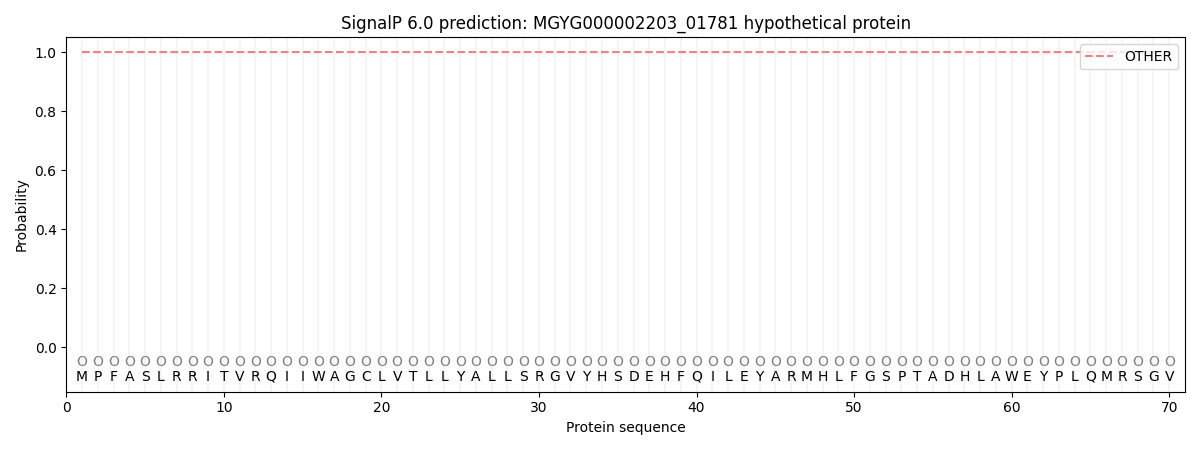
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
1.000041
0.000002
0.000000
0.000000
0.000000
0.000000
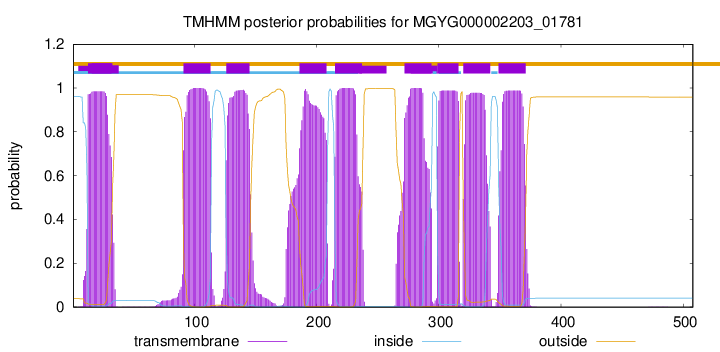
start
end
13
32
91
113
126
145
186
208
215
237
272
294
299
316
320
342
349
371