You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002203_00954
You are here: Home > Sequence: MGYG000002203_00954
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; | |||||||||||
| CAZyme ID | MGYG000002203_00954 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 90316; End: 93048 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 29 | 476 | 1.2e-64 | 0.5066489361702128 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10150 | PRK10150 | 1.55e-18 | 30 | 326 | 14 | 296 | beta-D-glucuronidase; Provisional |
| COG3250 | LacZ | 1.58e-14 | 95 | 465 | 66 | 444 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam00703 | Glyco_hydro_2 | 1.15e-06 | 199 | 306 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02837 | Glyco_hydro_2_N | 4.73e-06 | 31 | 163 | 4 | 135 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK10340 | ebgA | 4.23e-05 | 96 | 452 | 113 | 471 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK63994.1 | 0.0 | 1 | 906 | 1 | 905 |
| QPH40113.1 | 0.0 | 15 | 905 | 12 | 906 |
| AZI26708.1 | 0.0 | 24 | 907 | 24 | 908 |
| QIH37197.1 | 0.0 | 15 | 907 | 4 | 898 |
| VTR43024.1 | 0.0 | 14 | 905 | 14 | 904 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GM8_A | 4.64e-16 | 35 | 461 | 13 | 430 | ChainA, Glycoside hydrolase family 2, candidate beta-glycosidase [Phocaeicola vulgatus ATCC 8482] |
| 5T98_A | 6.57e-14 | 94 | 435 | 86 | 423 | Crystalstructure of BuGH2Awt [Bacteroides uniformis],5T98_B Crystal structure of BuGH2Awt [Bacteroides uniformis],5T99_A Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis],5T99_B Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis] |
| 6D8K_A | 1.05e-12 | 30 | 512 | 37 | 509 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
| 3K4A_A | 1.84e-12 | 20 | 211 | 7 | 196 | Crystalstructure of selenomethionine substituted E. coli beta-glucuronidase [Escherichia coli K-12],3K4A_B Crystal structure of selenomethionine substituted E. coli beta-glucuronidase [Escherichia coli K-12] |
| 6LEM_B | 3.17e-12 | 20 | 211 | 3 | 192 | ChainB, Beta-D-glucuronidase [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P05804 | 1.74e-11 | 20 | 211 | 5 | 194 | Beta-glucuronidase OS=Escherichia coli (strain K12) OX=83333 GN=uidA PE=1 SV=2 |
| A1SWB8 | 1.71e-06 | 18 | 429 | 38 | 460 | Beta-galactosidase OS=Psychromonas ingrahamii (strain 37) OX=357804 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
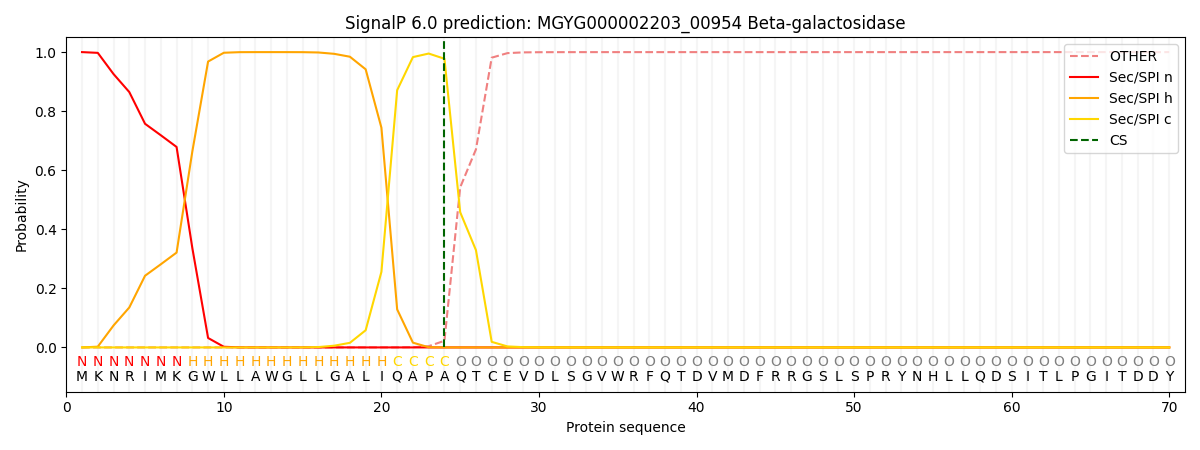
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000588 | 0.998209 | 0.000414 | 0.000244 | 0.000247 | 0.000254 |
