You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002166_00622
You are here: Home > Sequence: MGYG000002166_00622
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
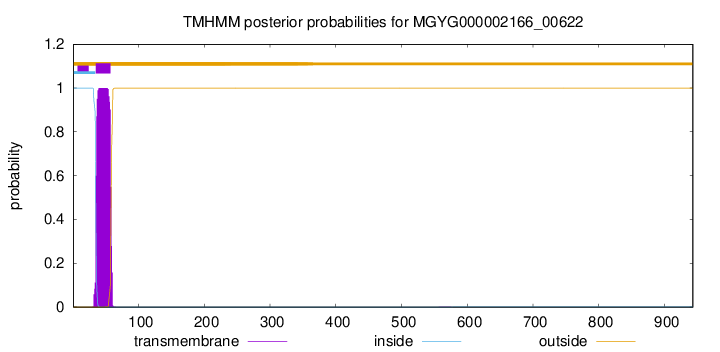
TMHMM annotations
Basic Information help
| Species | CAG-433 sp000433675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; CAG-433; CAG-433 sp000433675 | |||||||||||
| CAZyme ID | MGYG000002166_00622 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Monofunctional biosynthetic peptidoglycan transglycosylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11078; End: 13909 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 83 | 267 | 7.3e-54 | 0.9887005649717514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 5.15e-145 | 53 | 714 | 33 | 661 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 4.96e-98 | 53 | 726 | 22 | 796 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 1.75e-68 | 82 | 267 | 1 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| TIGR02071 | PBP_1b | 2.97e-67 | 92 | 620 | 145 | 656 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG4953 | PbpC | 1.52e-55 | 54 | 730 | 13 | 635 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNG61477.1 | 3.42e-160 | 50 | 728 | 53 | 715 |
| AKO91601.1 | 1.98e-157 | 51 | 728 | 53 | 742 |
| AXM90108.1 | 3.80e-156 | 51 | 703 | 46 | 687 |
| AKS38635.1 | 2.93e-155 | 51 | 703 | 46 | 687 |
| ACJ33515.1 | 2.93e-155 | 51 | 703 | 46 | 687 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DWK_A | 2.27e-99 | 78 | 664 | 10 | 602 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 2OLU_A | 1.96e-96 | 61 | 664 | 2 | 611 | StructuralInsight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Apoenzyme [Staphylococcus aureus],2OLV_A Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus],2OLV_B Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus] |
| 3ZG8_B | 1.65e-65 | 186 | 630 | 2 | 441 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 3ZG7_B | 6.13e-62 | 186 | 630 | 2 | 441 | CrystalStructure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the apo form [Listeria monocytogenes] |
| 2ZC5_B | 5.58e-48 | 303 | 666 | 11 | 379 | ChainB, Penicillin-binding protein 1A [unidentified],2ZC5_D Chain D, Penicillin-binding protein 1A [unidentified],2ZC6_B Chain B, Penicillin-binding protein 1A [unidentified],2ZC6_D Chain D, Penicillin-binding protein 1A [unidentified] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39793 | 8.32e-141 | 50 | 727 | 52 | 724 | Penicillin-binding protein 1A/1B OS=Bacillus subtilis (strain 168) OX=224308 GN=ponA PE=1 SV=1 |
| Q00573 | 3.80e-105 | 50 | 651 | 28 | 628 | Penicillin-binding protein 1A (Fragment) OS=Streptococcus oralis OX=1303 GN=ponA PE=3 SV=1 |
| Q04707 | 4.73e-101 | 50 | 666 | 29 | 642 | Penicillin-binding protein 1A OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=ponA PE=1 SV=2 |
| Q8DR59 | 6.58e-101 | 50 | 666 | 29 | 642 | Penicillin-binding protein 1A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=pbpA PE=1 SV=1 |
| P38050 | 2.62e-71 | 52 | 676 | 30 | 622 | Penicillin-binding protein 1F OS=Bacillus subtilis (strain 168) OX=224308 GN=pbpF PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
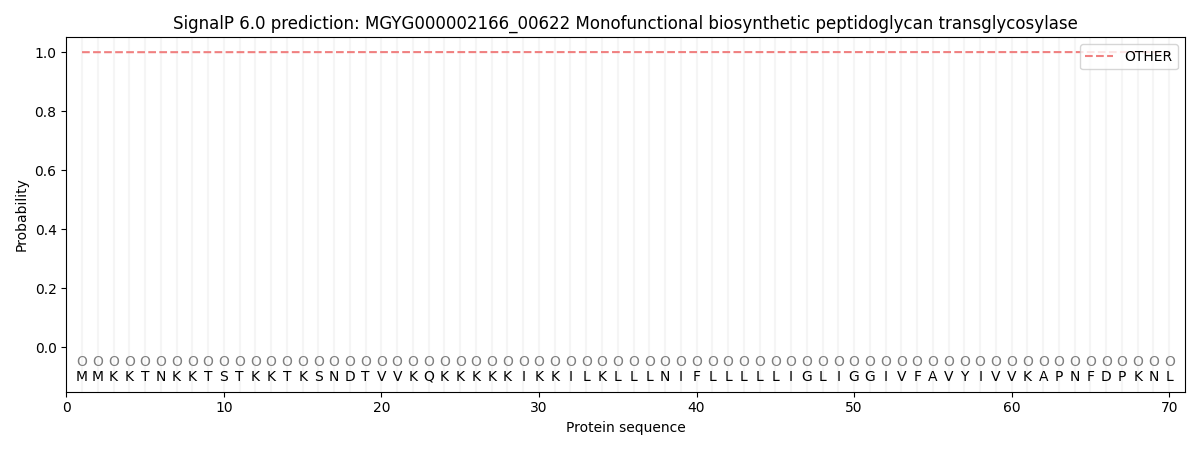
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999902 | 0.000121 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |