You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001940_01250
You are here: Home > Sequence: MGYG000001940_01250
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ruminococcus sp002438605 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; Ruminococcus sp002438605 | |||||||||||
| CAZyme ID | MGYG000001940_01250 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 351; End: 2204 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 50 | 367 | 2.7e-57 | 0.8888888888888888 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02773 | PLN02773 | 1.50e-32 | 50 | 353 | 14 | 263 | pectinesterase |
| PLN02682 | PLN02682 | 2.09e-31 | 21 | 337 | 43 | 311 | pectinesterase family protein |
| PLN02432 | PLN02432 | 2.11e-29 | 53 | 389 | 23 | 292 | putative pectinesterase |
| PLN02497 | PLN02497 | 2.33e-27 | 52 | 389 | 43 | 326 | probable pectinesterase |
| pfam01095 | Pectinesterase | 5.80e-27 | 53 | 375 | 12 | 275 | Pectinesterase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU21605.1 | 4.61e-139 | 41 | 414 | 365 | 742 |
| ADL49995.1 | 6.61e-114 | 7 | 393 | 5 | 404 |
| QAV09174.1 | 1.02e-109 | 41 | 412 | 509 | 891 |
| ADL52345.1 | 9.41e-106 | 41 | 445 | 508 | 913 |
| BAV13168.1 | 9.62e-106 | 41 | 445 | 524 | 929 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1GQ8_A | 1.55e-14 | 53 | 389 | 19 | 308 | Pectinmethylesterase from Carrot [Daucus carota] |
| 2NSP_A | 6.31e-14 | 130 | 390 | 69 | 339 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 5C1C_A | 1.76e-13 | 208 | 389 | 117 | 295 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 5C1E_A | 1.76e-13 | 208 | 389 | 117 | 295 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 3UW0_A | 2.41e-13 | 58 | 367 | 45 | 342 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FM79 | 1.29e-18 | 55 | 359 | 94 | 345 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
| Q9LVQ0 | 3.78e-17 | 53 | 353 | 17 | 263 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
| O04953 | 4.29e-15 | 209 | 339 | 101 | 232 | Putative pectinesterase 52 OS=Arabidopsis thaliana OX=3702 GN=PME52 PE=2 SV=2 |
| Q9ZQA3 | 7.45e-15 | 53 | 337 | 101 | 329 | Probable pectinesterase 15 OS=Arabidopsis thaliana OX=3702 GN=PME15 PE=2 SV=1 |
| A1DBT4 | 2.06e-14 | 208 | 367 | 142 | 296 | Probable pectinesterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pmeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
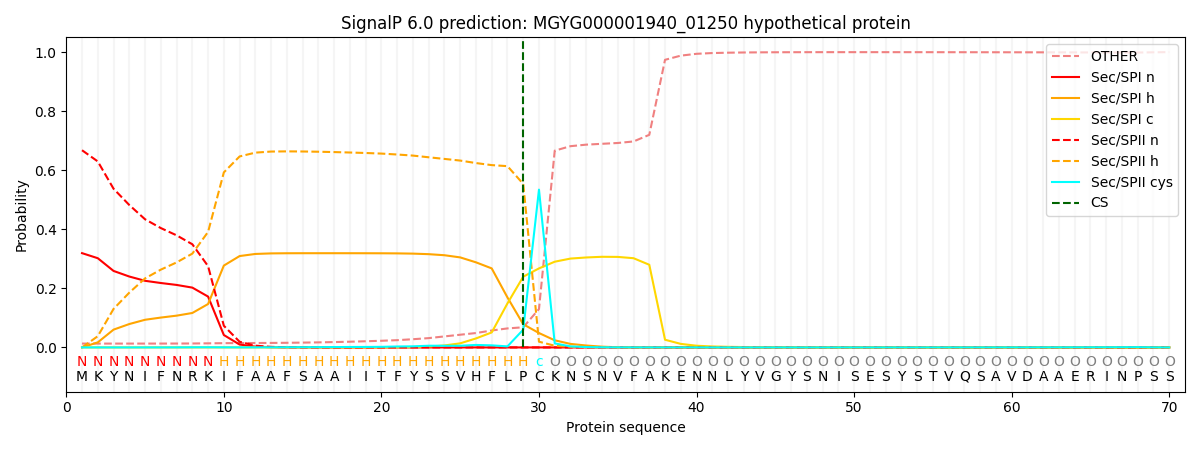
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.014850 | 0.312726 | 0.671829 | 0.000252 | 0.000176 | 0.000162 |
