You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001814_02414
You are here: Home > Sequence: MGYG000001814_02414
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | COE1 sp900753305 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; COE1; COE1 sp900753305 | |||||||||||
| CAZyme ID | MGYG000001814_02414 | |||||||||||
| CAZy Family | GH166 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17813; End: 18637 Strand: - | |||||||||||
Full Sequence Download help
| MKKIILKIFV IVLFVILLRG CFENTSKAME KYVVLIGEDS NVIDKITNID VLIIDAEYFS | 60 |
| KDEISHLKKN GVNEIYSYLN IGSIENFRPY YAEYEEYTLG EYENWPEEKW IDVSNPKWQE | 120 |
| FVASRVNDLV QKGVDGFFID NVDVYYMYQK ESIYDGIINI LKDIKNFDKK VIINGGDIFV | 180 |
| KKYLEKGKNK LIDGVNQENV YTMYDFSNDS YSKNAKDTRE YYTNYLDLVI QHGCTAYALE | 240 |
| YATNSVIQRE AEKYAKTHGY ICYVSDNIEL KLEK | 274 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH166 | 50 | 266 | 2.6e-60 | 0.9821428571428571 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03537 | Glyco_hydro_114 | 1.17e-24 | 49 | 145 | 17 | 112 | Glycoside-hydrolase family GH114. This family is recognized as a glycosyl-hydrolase family, number 114. It is endo-alpha-1,4-polygalactosaminidase, a rare enzyme. It is proposed to be TIM-barrel, the most common structure amongst the catalytic domains of glycosyl-hydrolases. |
| COG3868 | COG3868 | 1.02e-18 | 48 | 226 | 79 | 259 | Predicted glycosyl hydrolase, GH114 family [Carbohydrate transport and metabolism]. |
| COG2342 | COG2342 | 2.48e-18 | 26 | 273 | 22 | 288 | Endo alpha-1,4 polygalactosaminidase, GH114 family (was erroneously annotated as Cys-tRNA synthetase) [Carbohydrate transport and metabolism]. |
| TIGR01370 | TIGR01370 | 2.53e-09 | 55 | 265 | 79 | 303 | extracellular protein. Original assignment of this protein family as cysteinyl-tRNA synthetase is controversial, supported by but challenged by and by subsequent discovery of the actual mechanism for synthesizing Cys-tRNA in species where a direct Cys--tRNA ligase was not found. Lingering legacy annotations of members of this family probably should be removed. Evidence against the role includes a signal peptide. This family as been renamed "extracellular protein" to facilitate correction. Members of this family occur in Deinococcus radiodurans (bacterial) and Methanococcus jannaschii (archaeal). A number of homologous but more distantly related proteins are annotated as alpha-1,4 polygalactosaminidases. The function remains unknown. [Unknown function, General] |
| cd14745 | GH66 | 0.004 | 112 | 174 | 135 | 205 | Glycoside Hydrolase Family 66. Glycoside Hydrolase Family 66 contains proteins characterized as cycloisomaltooligosaccharide glucanotransferase (CITase) and dextranases from a variety of bacteria. CITase cyclizes part of a (1-6)-alpha-D-glucan (dextrans) chain by formation of a (1-6)-alpha-D-glucosidic bond. Dextranases catalyze the endohydrolysis of (1-6)-alpha-D-glucosidic linkages in dextran. Some members contain Carbohydrate Binding Module 35 (CBM35) domains, either C-terminal or inserted in the domain or both. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM04260.1 | 5.61e-112 | 35 | 272 | 1 | 238 |
| QHI71547.1 | 2.65e-73 | 9 | 270 | 4 | 271 |
| AOZ95737.1 | 2.65e-71 | 2 | 271 | 8 | 286 |
| ASS41767.1 | 4.69e-67 | 11 | 272 | 4 | 268 |
| ADU22174.1 | 4.11e-66 | 20 | 270 | 23 | 274 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
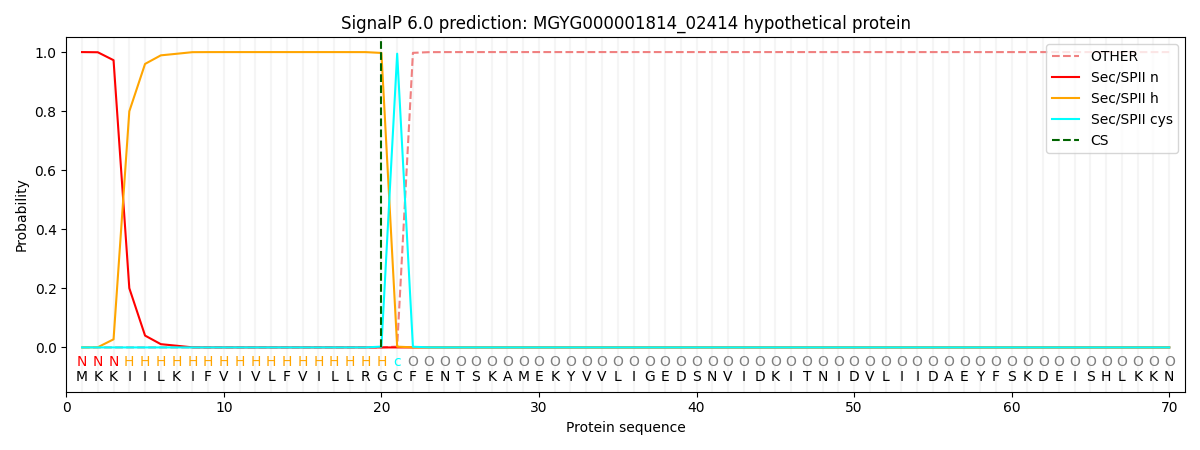
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000048 | 0.000000 | 0.000000 | 0.000000 |

