You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001783_00693
You are here: Home > Sequence: MGYG000001783_00693
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900551065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900551065 | |||||||||||
| CAZyme ID | MGYG000001783_00693 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18490; End: 20163 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 206 | 507 | 4e-91 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.71e-48 | 203 | 507 | 13 | 269 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.44e-18 | 151 | 553 | 21 | 407 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam13205 | Big_5 | 1.48e-09 | 63 | 157 | 6 | 106 | Bacterial Ig-like domain. |
| pfam04234 | CopC | 0.002 | 62 | 118 | 2 | 68 | CopC domain. CopC is a bacterial blue copper protein that binds 1 atom of copper per protein molecule. Along with CopA, CopC mediates copper resistance by sequestration of copper in the periplasm. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIK55671.1 | 3.05e-103 | 162 | 537 | 128 | 478 |
| QIK61079.1 | 1.70e-102 | 162 | 537 | 128 | 478 |
| ADQ18612.1 | 3.18e-92 | 168 | 552 | 194 | 552 |
| BAF57333.1 | 3.38e-91 | 171 | 547 | 20 | 366 |
| ASB50110.1 | 2.87e-88 | 161 | 551 | 215 | 573 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OYC_A | 1.07e-74 | 166 | 551 | 36 | 393 | GH5endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYC_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107] |
| 6HA9_A | 8.23e-74 | 166 | 551 | 36 | 393 | Structureof an endo-Xyloglucanase from Cellvibrio japonicus complexed with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HA9_B Structure of an endo-Xyloglucanase from Cellvibrio japonicus complexed with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HAA_A Structure of a covalent complex of endo-Xyloglucanase from Cellvibrio japonicus after reacting with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107],6HAA_B Structure of a covalent complex of endo-Xyloglucanase from Cellvibrio japonicus after reacting with XXXG(2F)-beta-DNP [Cellvibrio japonicus Ueda107] |
| 1EDG_A | 2.57e-64 | 158 | 530 | 4 | 357 | SingleCrystal Structure Determination Of The Catalytic Domain Of Celcca Carried Out At 15 Degree C [Ruminiclostridium cellulolyticum H10] |
| 3ZMR_A | 2.20e-61 | 166 | 515 | 105 | 447 | Bacteroidesovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide [Bacteroides ovatus],3ZMR_B Bacteroides ovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide [Bacteroides ovatus] |
| 6XSU_A | 8.35e-61 | 175 | 549 | 13 | 351 | GH5-4broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens],6XSU_B GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P17901 | 1.58e-62 | 158 | 530 | 29 | 382 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| A7LXT7 | 3.03e-61 | 39 | 515 | 23 | 482 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH5A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02653 PE=1 SV=1 |
| O08342 | 8.35e-57 | 181 | 552 | 46 | 400 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P23660 | 2.45e-56 | 175 | 552 | 27 | 364 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P54937 | 1.20e-55 | 153 | 556 | 17 | 383 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
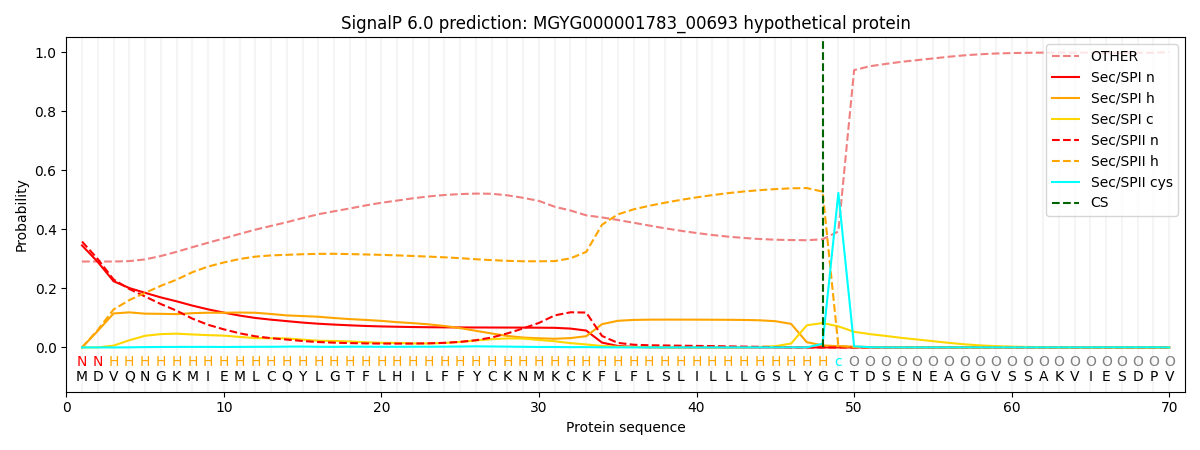
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.293250 | 0.207188 | 0.492887 | 0.000511 | 0.005558 | 0.000603 |
