You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001732_01184
You are here: Home > Sequence: MGYG000001732_01184
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
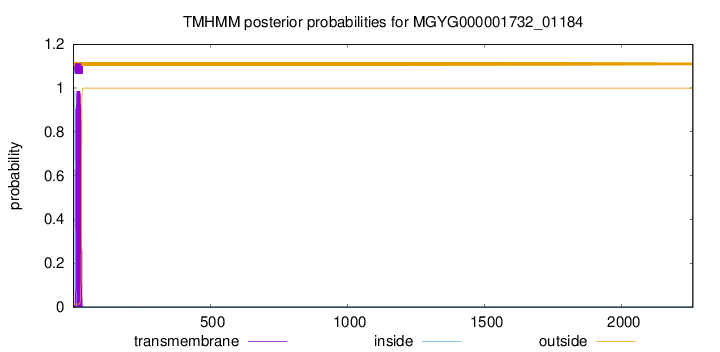
TMHMM annotations
Basic Information help
| Species | HGM11530 sp900751685 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; HGM11530; HGM11530 sp900751685 | |||||||||||
| CAZyme ID | MGYG000001732_01184 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20946; End: 27725 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 1198 | 2195 | 5.1e-80 | 0.8976063829787234 |
| CE1 | 546 | 759 | 8.1e-31 | 0.8942731277533039 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.52e-40 | 1247 | 1746 | 47 | 483 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| COG4099 | COG4099 | 4.65e-31 | 505 | 783 | 134 | 387 | Predicted peptidase [General function prediction only]. |
| PRK10150 | PRK10150 | 3.67e-29 | 1269 | 1721 | 68 | 447 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.08e-27 | 1270 | 1721 | 113 | 474 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02836 | Glyco_hydro_2_C | 1.61e-18 | 1547 | 1781 | 4 | 213 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI62299.1 | 4.60e-132 | 1205 | 2252 | 23 | 989 |
| QFK71524.1 | 4.19e-98 | 1208 | 1959 | 5 | 639 |
| AIQ56230.1 | 1.31e-95 | 1205 | 1950 | 2 | 632 |
| CEI73305.1 | 2.34e-95 | 1208 | 1957 | 5 | 638 |
| QIZ08201.1 | 1.92e-92 | 1208 | 1957 | 5 | 639 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CMG_A | 2.80e-81 | 1245 | 1957 | 28 | 639 | Crystalstructure of putative beta-galactosidase from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 5Z1A_A | 3.78e-81 | 1245 | 1957 | 47 | 658 | Thecrystal structure of Bacteroides fragilis beta-glucuronidase in complex with uronic isofagomine [Bacteroides fragilis NCTC 9343] |
| 6MVG_A | 1.35e-68 | 1209 | 1921 | 29 | 620 | Crystalstructure of FMN-binding beta-glucuronidase from Ruminococcus gnavus [[Ruminococcus] gnavus],6MVG_B Crystal structure of FMN-binding beta-glucuronidase from Ruminococcus gnavus [[Ruminococcus] gnavus],6MVG_C Crystal structure of FMN-binding beta-glucuronidase from Ruminococcus gnavus [[Ruminococcus] gnavus] |
| 3FN9_A | 1.66e-68 | 1242 | 1936 | 36 | 636 | Crystalstructure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_B Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_C Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_D Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 6MVH_A | 1.25e-60 | 1200 | 1921 | 20 | 620 | Crystalstructure of FMN-binding beta-glucuronidase from Roseburia hominis [Roseburia hominis],6MVH_B Crystal structure of FMN-binding beta-glucuronidase from Roseburia hominis [Roseburia hominis],6MVH_C Crystal structure of FMN-binding beta-glucuronidase from Roseburia hominis [Roseburia hominis],6MVH_D Crystal structure of FMN-binding beta-glucuronidase from Roseburia hominis [Roseburia hominis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPJ7 | 1.47e-50 | 1200 | 1921 | 46 | 673 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| A7LXS9 | 2.33e-32 | 1211 | 1725 | 48 | 503 | Beta-galactosidase BoGH2A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02645 PE=1 SV=1 |
| P77989 | 1.11e-28 | 1208 | 1667 | 6 | 382 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| P26257 | 2.65e-23 | 1239 | 1719 | 27 | 413 | Beta-galactosidase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=lacZ PE=1 SV=1 |
| Q6LL68 | 1.54e-15 | 1486 | 1721 | 278 | 481 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
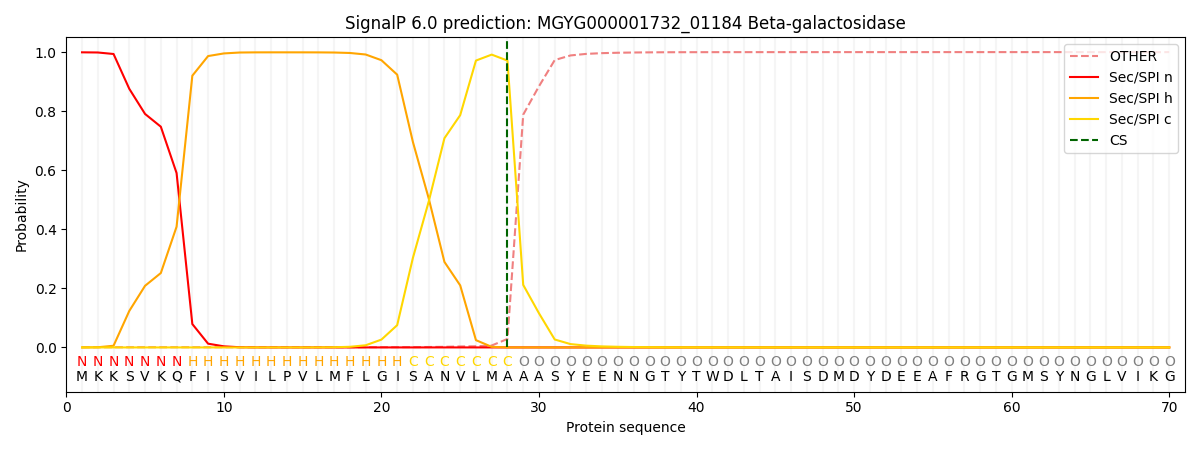
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001821 | 0.997157 | 0.000283 | 0.000249 | 0.000223 | 0.000211 |