You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001684_01353
You are here: Home > Sequence: MGYG000001684_01353
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
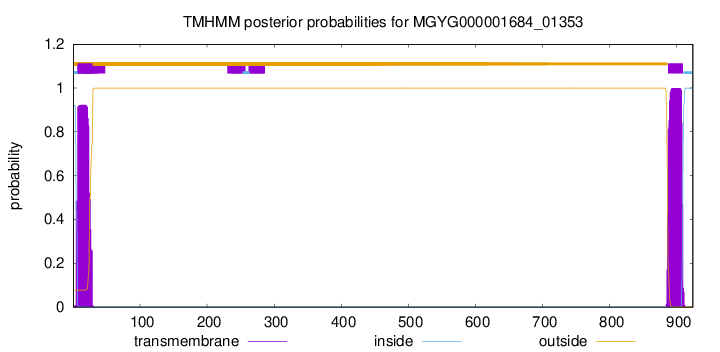
TMHMM annotations
Basic Information help
| Species | DONG20-135 sp009831105 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; DONG20-135; DONG20-135 sp009831105 | |||||||||||
| CAZyme ID | MGYG000001684_01353 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 600; End: 3374 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 304 | 417 | 7.2e-17 | 0.9453125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 3.46e-14 | 236 | 422 | 59 | 233 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam01832 | Glucosaminidase | 3.70e-08 | 304 | 374 | 4 | 81 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| pfam07523 | Big_3 | 1.97e-05 | 597 | 661 | 3 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| smart00047 | LYZ2 | 3.03e-05 | 292 | 422 | 4 | 144 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| cd12087 | TM_EGFR-like | 0.005 | 888 | 919 | 6 | 38 | Transmembrane domain of the Epidermal Growth Factor Receptor family of Protein Tyrosine Kinases. PTKs catalyze the transfer of the gamma-phosphoryl group from ATP to tyrosine (tyr) residues in protein substrates. EGFR (HER, ErbB) subfamily members include EGFR (HER1, ErbB1), HER2 (ErbB2), HER3 (ErbB3), HER4 (ErbB4), and similar proteins. They are receptor PTKs (RTKs) containing an extracellular EGF-related ligand-binding region, a transmembrane (TM) helix, and a cytoplasmic region with a tyr kinase domain and a regulatory C-terminal tail. They are activated by ligand-induced dimerization, resulting in the phosphorylation of tyr residues in the C-terminal tail, which serve as binding sites for downstream signaling molecules. Collectively, they can recognize a variety of ligands including EGF, TGFalpha, and neuregulins, among others. All four subfamily members can form homo- or heterodimers. HER3 contains an impaired kinase domain and depends on its heterodimerization partner for activation. EGFR subfamily members are involved in signaling pathways leading to a broad range of cellular responses including cell proliferation, differentiation, migration, growth inhibition, and apoptosis. The TM domain not only serves as a membrane anchor, but also plays an important role in receptor dimerization and optimal activation. Mutations in the TM domain of EGFR family RTKs have been associated with increased breast cancer risk. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSI25075.1 | 2.99e-210 | 2 | 879 | 4 | 875 |
| QQR25465.1 | 1.58e-204 | 2 | 879 | 4 | 875 |
| QIX10256.1 | 1.58e-204 | 2 | 879 | 4 | 875 |
| ANU70664.1 | 1.58e-204 | 2 | 879 | 4 | 875 |
| ASU20850.1 | 1.58e-204 | 2 | 879 | 4 | 875 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 5.47e-19 | 187 | 425 | 69 | 290 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 1.62e-18 | 194 | 425 | 447 | 658 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 1.77e-18 | 194 | 425 | 491 | 702 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000793 | 0.751118 | 0.247373 | 0.000250 | 0.000238 | 0.000207 |