You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001591_04202
You are here: Home > Sequence: MGYG000001591_04202
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
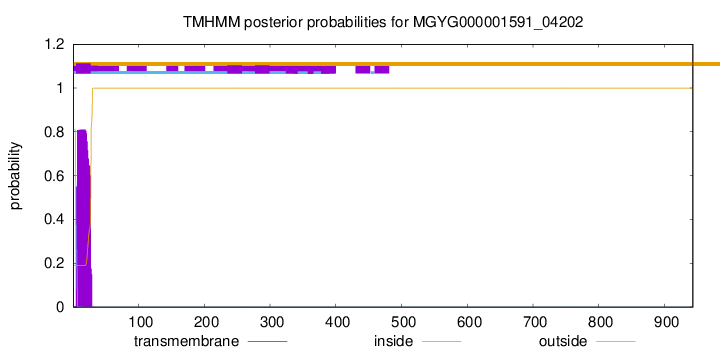
TMHMM annotations
Basic Information help
| Species | Parabacteroides timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides timonensis | |||||||||||
| CAZyme ID | MGYG000001591_04202 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10742; End: 13573 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 130 | 201 | 4.1e-19 | 0.5298507462686567 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 1.79e-37 | 27 | 202 | 3 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 8.75e-28 | 27 | 202 | 5 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT48267.1 | 0.0 | 27 | 942 | 25 | 938 |
| AWW33261.1 | 0.0 | 15 | 942 | 11 | 925 |
| QUR47740.1 | 0.0 | 1 | 942 | 1 | 935 |
| QUT51561.1 | 0.0 | 1 | 942 | 1 | 935 |
| QCY57605.1 | 0.0 | 1 | 942 | 1 | 935 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JS4_A | 1.83e-09 | 125 | 318 | 676 | 859 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 2VMH_A | 1.59e-06 | 27 | 198 | 9 | 147 | Thestructure of CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 2VMI_A | 1.59e-06 | 27 | 198 | 9 | 147 | Thestructure of seleno-methionine labelled CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 2VMG_A | 1.78e-06 | 27 | 198 | 15 | 153 | Thestructure of CBM51 from Clostridium perfringens GH95 in complex with methyl-galactose [Clostridium perfringens] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
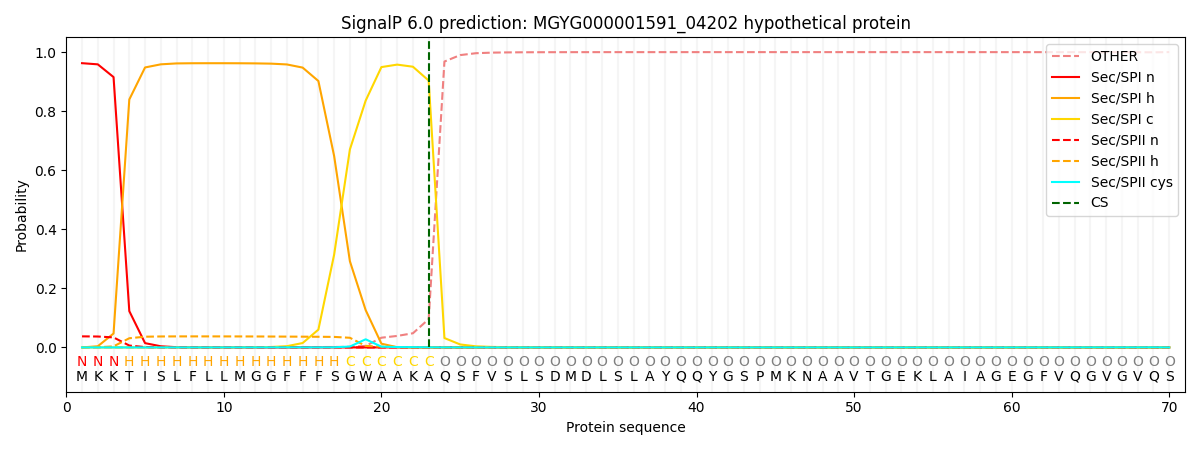
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000994 | 0.957871 | 0.040221 | 0.000310 | 0.000294 | 0.000259 |