You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001576_01011
You are here: Home > Sequence: MGYG000001576_01011
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
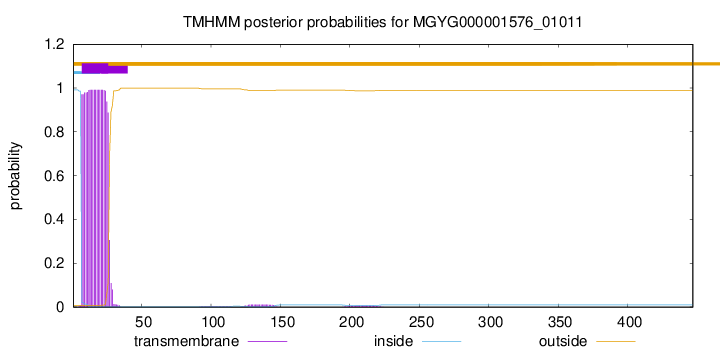
TMHMM annotations
Basic Information help
| Species | Dysosmobacter sp014297375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Dysosmobacter; Dysosmobacter sp014297375 | |||||||||||
| CAZyme ID | MGYG000001576_01011 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 94602; End: 95945 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02252 | nylC_like | 2.80e-92 | 38 | 347 | 3 | 259 | nylC-like family; composed of proteins with similarity to Flavobacterium endo-type 6-aminohexanoate-oligomer hydrolase (EIII), the product of the nylon oligomer degradation gene, nylC. EIII is an amide hydrolase that catalyzes the degradation of highly-polymerized 6-aminohexanoate oligomers. Together with other nylon degradation enzymes, such as 6-aminohexanoate cyclic dimer hydrolase (EI) and 6-aminohexanoate dimer hydrolase (EII), EIII plays a role in the detoxification and biological removal of the synthetic by-products of nylon manufacture. EIII shows sequence similarity to L-aminopeptidase D-amidase/D-esterase (DmpA), an aminopeptidase that releases N-terminal D and L amino acids from peptide substrates. Like DmpA, EIII undergoes autocatalytic cleavage in front of a nucleophile to form a heterodimer. DmpA shows similarity in catalytic mechanism to N-terminal nucleophile (Ntn) hydrolases, which are enzymes that catalyze the cleavage of amide bonds through the nucleophilic attack of the side chain of an N-terminal serine, threonine, or cysteine. |
| COG3191 | DmpA | 7.00e-68 | 38 | 363 | 19 | 339 | L-aminopeptidase/D-esterase [Amino acid transport and metabolism, Secondary metabolites biosynthesis, transport and catabolism]. |
| pfam03576 | Peptidase_S58 | 4.57e-46 | 38 | 356 | 5 | 309 | Peptidase family S58. |
| cd00118 | LysM | 3.42e-16 | 401 | 445 | 1 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
| pfam01476 | LysM | 1.76e-15 | 403 | 446 | 1 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCK83709.1 | 3.07e-301 | 1 | 447 | 1 | 447 |
| QHZ47012.1 | 2.77e-10 | 399 | 445 | 214 | 260 |
| CDM70433.1 | 6.98e-10 | 374 | 446 | 194 | 266 |
| ASB88452.1 | 8.99e-10 | 385 | 445 | 203 | 261 |
| QFG73306.1 | 1.27e-09 | 388 | 445 | 408 | 464 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3TM1_A | 4.69e-54 | 38 | 369 | 57 | 405 | Crystalstructure of mature ThnT, a pantetheine hydrolase [Streptomyces cattleya],3TM1_B Crystal structure of mature ThnT, a pantetheine hydrolase [Streptomyces cattleya],3TM2_A Crystal structure of mature ThnT with a covalently bound product mimic [Streptomyces cattleya],3TM2_B Crystal structure of mature ThnT with a covalently bound product mimic [Streptomyces cattleya] |
| 4IHE_A | 2.50e-53 | 38 | 369 | 57 | 405 | CrystalStructure of Uncleaved ThnT T282A [Streptomyces cattleya],4IHE_B Crystal Structure of Uncleaved ThnT T282A [Streptomyces cattleya] |
| 3S3U_A | 3.50e-53 | 38 | 369 | 57 | 405 | CrystalStructure of Uncleaved ThnT T282C [Streptomyces cattleya],3S3U_B Crystal Structure of Uncleaved ThnT T282C [Streptomyces cattleya],4IHD_A Crystal Structure of Uncleaved ThnT T282C, derivatized at the active site with EtHg [Streptomyces cattleya],4IHD_B Crystal Structure of Uncleaved ThnT T282C, derivatized at the active site with EtHg [Streptomyces cattleya] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P64812 | 5.01e-50 | 38 | 358 | 6 | 332 | Uncharacterized aminopeptidase Mb1368 OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=BQ2027_MB1368 PE=3 SV=1 |
| P9WM22 | 5.01e-50 | 38 | 358 | 6 | 332 | Uncharacterized aminopeptidase MT1375 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT1375 PE=3 SV=1 |
| P9WM23 | 5.01e-50 | 38 | 358 | 6 | 332 | Uncharacterized aminopeptidase Rv1333 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=Rv1333 PE=1 SV=1 |
| P53425 | 2.15e-49 | 38 | 362 | 13 | 347 | Uncharacterized aminopeptidase ML1167 OS=Mycobacterium leprae (strain TN) OX=272631 GN=ML1167 PE=3 SV=1 |
| Q5HRU2 | 6.19e-07 | 396 | 444 | 79 | 126 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
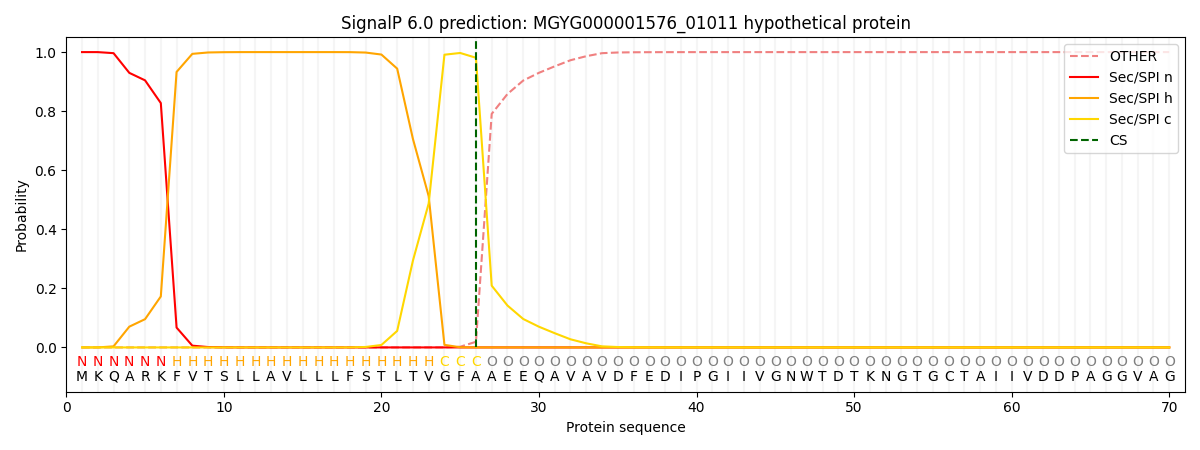
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000202 | 0.999190 | 0.000165 | 0.000160 | 0.000145 | 0.000134 |