You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001542_04012
You are here: Home > Sequence: MGYG000001542_04012
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A sp900069005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A sp900069005 | |||||||||||
| CAZyme ID | MGYG000001542_04012 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35859; End: 37754 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 47 | 360 | 1.6e-138 | 0.9967741935483871 |
| CBM3 | 495 | 561 | 4.8e-20 | 0.8181818181818182 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2730 | BglC | 4.96e-55 | 40 | 379 | 32 | 389 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam00150 | Cellulase | 9.87e-52 | 46 | 355 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| pfam00942 | CBM_3 | 6.49e-20 | 494 | 566 | 1 | 82 | Cellulose binding domain. |
| smart01067 | CBM_3 | 4.51e-18 | 494 | 567 | 1 | 83 | Cellulose binding domain. |
| cd00063 | FN3 | 3.12e-10 | 394 | 479 | 1 | 92 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZS14931.1 | 0.0 | 1 | 631 | 1 | 629 |
| QJD87157.1 | 0.0 | 33 | 631 | 34 | 632 |
| QKS44641.1 | 3.08e-311 | 33 | 631 | 34 | 729 |
| ASA23029.1 | 3.08e-249 | 33 | 631 | 28 | 557 |
| AIQ39244.1 | 3.19e-248 | 30 | 631 | 30 | 554 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1VRX_A | 8.94e-143 | 33 | 380 | 1 | 350 | ChainA, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus],1VRX_B Chain B, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus] |
| 1ECE_A | 2.53e-142 | 33 | 380 | 1 | 350 | AcidothermusCellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus],1ECE_B Acidothermus Cellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus] |
| 4TUF_A | 1.61e-104 | 37 | 377 | 1 | 340 | Catalyticdomain of the major endoglucanase from Xanthomonas campestris pv. campestris [Xanthomonas campestris pv. campestris str. ATCC 33913],4TUF_B Catalytic domain of the major endoglucanase from Xanthomonas campestris pv. campestris [Xanthomonas campestris pv. campestris str. ATCC 33913],4TUF_C Catalytic domain of the major endoglucanase from Xanthomonas campestris pv. campestris [Xanthomonas campestris pv. campestris str. ATCC 33913],4TUF_D Catalytic domain of the major endoglucanase from Xanthomonas campestris pv. campestris [Xanthomonas campestris pv. campestris str. ATCC 33913] |
| 3W6M_A | 9.29e-100 | 37 | 381 | 7 | 376 | Contributionof disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
| 3VVG_A | 1.31e-99 | 37 | 381 | 7 | 376 | TheCrystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_B The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_C The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3W6L_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23548 | 1.15e-209 | 31 | 384 | 32 | 385 | Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
| P54583 | 1.18e-138 | 33 | 380 | 42 | 391 | Endoglucanase E1 OS=Acidothermus cellulolyticus (strain ATCC 43068 / DSM 8971 / 11B) OX=351607 GN=Acel_0614 PE=1 SV=1 |
| P19487 | 1.69e-102 | 31 | 377 | 20 | 365 | Major extracellular endoglucanase OS=Xanthomonas campestris pv. campestris (strain ATCC 33913 / DSM 3586 / NCPPB 528 / LMG 568 / P 25) OX=190485 GN=engXCA PE=1 SV=2 |
| P50400 | 1.78e-82 | 30 | 439 | 38 | 503 | Endoglucanase D OS=Cellulomonas fimi OX=1708 GN=cenD PE=3 SV=1 |
| P10474 | 8.11e-75 | 36 | 377 | 628 | 993 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
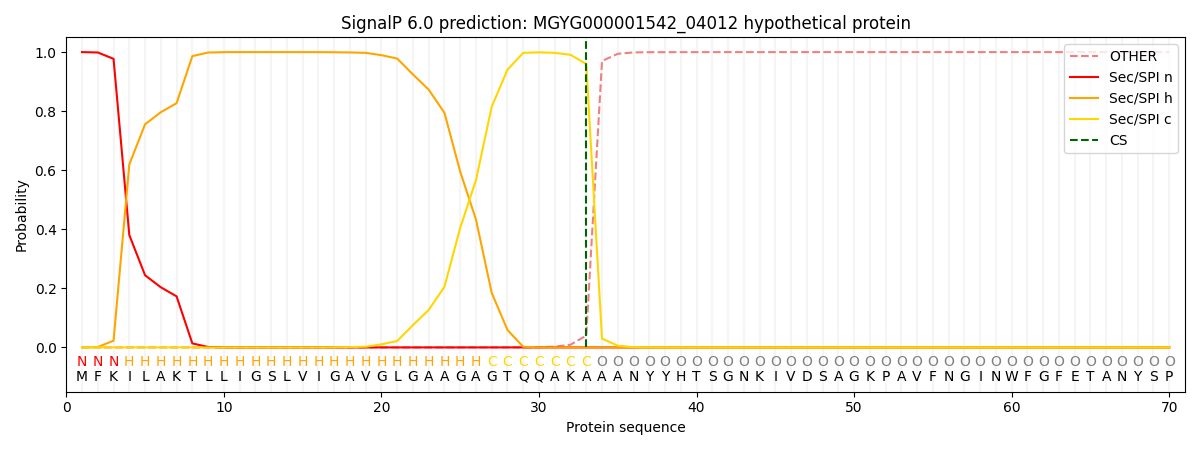
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000372 | 0.998898 | 0.000197 | 0.000187 | 0.000169 | 0.000159 |
