You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001537_03655
You are here: Home > Sequence: MGYG000001537_03655
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
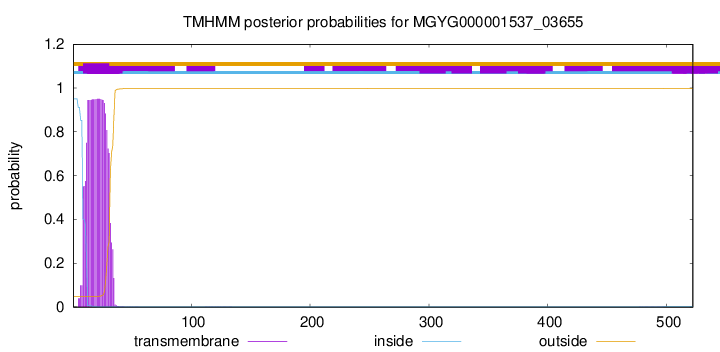
TMHMM annotations
Basic Information help
| Species | Cellulomonas timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas timonensis | |||||||||||
| CAZyme ID | MGYG000001537_03655 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1015771; End: 1017339 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 104 | 285 | 2.4e-73 | 0.989247311827957 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 6.86e-51 | 37 | 359 | 32 | 343 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 4.06e-47 | 105 | 287 | 7 | 190 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 4.55e-25 | 114 | 283 | 34 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVI66103.1 | 1.23e-266 | 1 | 521 | 1 | 519 |
| AJP05029.1 | 2.87e-266 | 1 | 521 | 1 | 523 |
| CQR60281.1 | 2.79e-263 | 54 | 521 | 39 | 510 |
| QMU17794.1 | 2.00e-262 | 1 | 521 | 1 | 516 |
| ALC19118.1 | 2.00e-262 | 1 | 521 | 1 | 516 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VMV_A | 1.44e-50 | 54 | 356 | 19 | 321 | Crystalstructure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
| 2QXZ_A | 7.13e-35 | 108 | 361 | 66 | 328 | ChainA, pectate lyase II [Xanthomonas campestris pv. campestris],2QXZ_B Chain B, pectate lyase II [Xanthomonas campestris pv. campestris] |
| 2QY1_A | 7.13e-35 | 108 | 361 | 66 | 328 | ChainA, Pectate lyase II [Xanthomonas campestris pv. campestris],2QY1_B Chain B, Pectate lyase II [Xanthomonas campestris pv. campestris] |
| 2QX3_A | 1.75e-33 | 108 | 361 | 66 | 328 | Structureof pectate lyase II from Xanthomonas campestris pv. campestris str. ATCC 33913 [Xanthomonas campestris pv. campestris],2QX3_B Structure of pectate lyase II from Xanthomonas campestris pv. campestris str. ATCC 33913 [Xanthomonas campestris pv. campestris] |
| 1AIR_A | 3.81e-33 | 108 | 361 | 82 | 344 | ChainA, PECTATE LYASE C [Dickeya chrysanthemi],1O88_A Chain A, Pectate Lyase C [Dickeya chrysanthemi],1O8D_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8E_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8F_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8G_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8H_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8I_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8J_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8K_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8L_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8M_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1PLU_A Chain A, Protein (pectate Lyase C) [Dickeya chrysanthemi],2PEC_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1B6T1 | 2.26e-63 | 54 | 356 | 59 | 335 | Pectate trisaccharide-lyase OS=Bacillus sp. OX=1409 GN=pel PE=1 SV=1 |
| Q65DC2 | 2.26e-63 | 54 | 356 | 59 | 335 | Pectate trisaccharide-lyase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=BLi04129 PE=3 SV=1 |
| Q8GCB2 | 2.26e-63 | 54 | 356 | 59 | 335 | Pectate trisaccharide-lyase OS=Bacillus licheniformis OX=1402 GN=pelA PE=1 SV=1 |
| P04959 | 3.96e-35 | 108 | 361 | 103 | 366 | Pectate lyase B OS=Dickeya chrysanthemi OX=556 GN=pelB PE=3 SV=1 |
| P0C1C2 | 3.09e-32 | 108 | 363 | 103 | 367 | Pectate lyase 3 OS=Pectobacterium carotovorum OX=554 GN=pel3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002241 | 0.992056 | 0.000371 | 0.004478 | 0.000473 | 0.000318 |