You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001489_01368
You are here: Home > Sequence: MGYG000001489_01368
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides goldsteinii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides goldsteinii | |||||||||||
| CAZyme ID | MGYG000001489_01368 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1770478; End: 1772592 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 394 | 604 | 2.6e-19 | 0.5584795321637427 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 1.71e-22 | 359 | 693 | 17 | 333 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| COG1957 | URH1 | 3.39e-16 | 38 | 212 | 3 | 174 | Inosine-uridine nucleoside N-ribohydrolase [Nucleotide transport and metabolism]. |
| cd02651 | nuc_hydro_IU_UC_XIUA | 2.40e-15 | 40 | 324 | 2 | 298 | nuc_hydro_IU_UC_XIUA: inosine-uridine preferring, xanthosine-inosine-uridine-adenosine-preferring and, uridine-cytidine preferring nucleoside hydrolases. Nucleoside hydrolases cleave the N-glycosidic bond in nucleosides generating ribose and the respective base. These enzymes vary in their substrate specificity. This group contains proteins similar to nucleoside hydrolases which hydrolyze both pyrimidine and purine ribonucleosides: the inosine-uridine preferring nucleoside hydrolase from Crithidia fasciculata, the inosine-uridine-xanthosine preferring nucleoside hydrolase RihC from Escherichia coli and the xanthosine-inosine-uridine-adenosine-preferring nucleoside hydrolase RihC from Salmonella enterica serovar Typhimurium. This group also contains proteins similar to the pyrimidine-specific uridine-cytidine preferring nucleoside hydrolases URH1 from Saccharomyces cerevisiae, E. coli RihA and E. coli RihB. E. coli RihA is equally efficient with uridine and cytidine, E. coli RihB prefers cytidine over uridine. S. cerevisiae URH1 prefers uridine over cytidine. |
| cd00455 | nuc_hydro | 7.77e-15 | 40 | 302 | 1 | 261 | nuc_hydro: Nucleoside hydrolases. Nucleoside hydrolases cleave the N-glycosidic bond in nucleosides generating ribose and the respective base. These enzymes vary in their substrate specificity. This group contains eukaryotic, bacterial and archeal proteins similar to the inosine-uridine preferring nucleoside hydrolase from Crithidia fasciculata, the xanthosine-inosine-uridine-adenosine-preferring nucleoside hydrolase RihC from Salmonella enterica serovar Typhimurium, the purine-specific inosine-adenosine-guanosine-preferring nucleoside hydrolase from Trypanosoma vivax and, pyrimidine-specific uridine-cytidine preferring nucleoside hydrolases such as URH1 from Saccharomyces cerevisiae, RihA and RihB from Escherichia coli. Nucleoside hydrolases are of interest as a target for antiprotozoan drugs as, no nucleoside hydrolase activity or genes encoding these enzymes have been detected in humans and, parasitic protozoans lack de novo purine synthesis relying on nucleoside hydrolase to scavenge purine and/or pyrimidines from the environment. |
| pfam01156 | IU_nuc_hydro | 4.25e-14 | 38 | 322 | 1 | 255 | Inosine-uridine preferring nucleoside hydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCM58084.1 | 3.26e-188 | 341 | 698 | 37 | 393 |
| BBD46309.1 | 3.44e-185 | 340 | 701 | 35 | 395 |
| ACT93133.1 | 2.23e-171 | 320 | 700 | 12 | 398 |
| SOE23805.1 | 2.19e-170 | 340 | 700 | 25 | 382 |
| QGY48039.1 | 1.67e-167 | 342 | 700 | 27 | 384 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5HX0_A | 9.46e-170 | 320 | 700 | 15 | 401 | ChainA, Uncharacterized protein Dfer_1899 [Dyadobacter fermentans DSM 18053],5HX0_B Chain B, Uncharacterized protein Dfer_1899 [Dyadobacter fermentans DSM 18053] |
| 4KPN_A | 3.46e-10 | 38 | 222 | 30 | 215 | Plantnucleoside hydrolase - PpNRh1 enzyme [Physcomitrium patens],4KPN_B Plant nucleoside hydrolase - PpNRh1 enzyme [Physcomitrium patens],4KPN_C Plant nucleoside hydrolase - PpNRh1 enzyme [Physcomitrium patens],4KPN_D Plant nucleoside hydrolase - PpNRh1 enzyme [Physcomitrium patens],4KPN_E Plant nucleoside hydrolase - PpNRh1 enzyme [Physcomitrium patens],4KPN_F Plant nucleoside hydrolase - PpNRh1 enzyme [Physcomitrium patens],4KPN_G Plant nucleoside hydrolase - PpNRh1 enzyme [Physcomitrium patens],4KPN_H Plant nucleoside hydrolase - PpNRh1 enzyme [Physcomitrium patens] |
| 6BA1_A | 2.13e-06 | 37 | 216 | 4 | 184 | Purine-PreferringRibonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_B Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_C Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_D Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_E Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_F Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_G Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_H Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_I Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_J Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_K Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_L Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_M Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_N Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_O Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_P Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_Q Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA1_R Purine-Preferring Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A] |
| 6BA0_A | 2.75e-06 | 40 | 254 | 7 | 213 | Pyrimidine-specificRibonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA0_B Pyrimidine-specific Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA0_C Pyrimidine-specific Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A],6BA0_D Pyrimidine-specific Ribonucleoside Hydrolase from Gardnerella vaginalis [Gardnerella vaginalis 315-A] |
| 3G5I_A | 4.80e-06 | 40 | 253 | 6 | 210 | CrystalStructure of the E.coli RihA pyrimidine nucleosidase bound to a iminoribitol-based inhibitor [Escherichia coli K-12],3G5I_B Crystal Structure of the E.coli RihA pyrimidine nucleosidase bound to a iminoribitol-based inhibitor [Escherichia coli K-12],3G5I_C Crystal Structure of the E.coli RihA pyrimidine nucleosidase bound to a iminoribitol-based inhibitor [Escherichia coli K-12],3G5I_D Crystal Structure of the E.coli RihA pyrimidine nucleosidase bound to a iminoribitol-based inhibitor [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8LAC4 | 6.84e-09 | 38 | 220 | 7 | 190 | Probable uridine nucleosidase 2 OS=Arabidopsis thaliana OX=3702 GN=URH2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
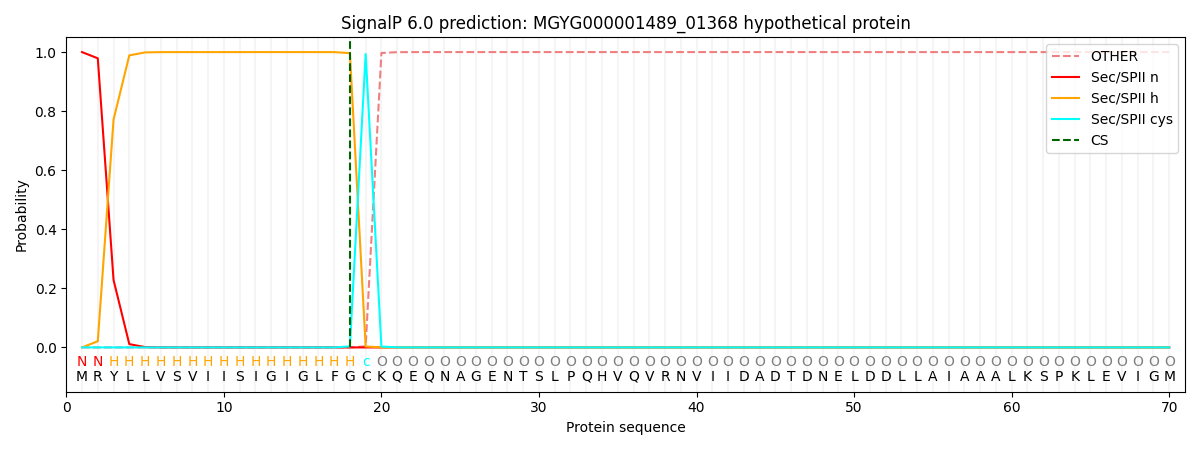
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000065 | 0.000000 | 0.000000 | 0.000000 |
