You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001443_03084
You are here: Home > Sequence: MGYG000001443_03084
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
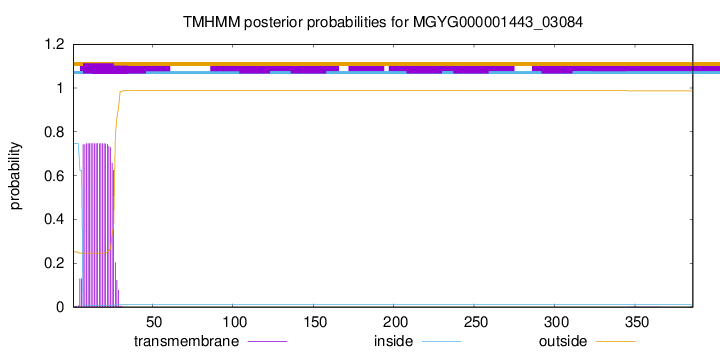
TMHMM annotations
Basic Information help
| Species | Streptomyces albus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces albus | |||||||||||
| CAZyme ID | MGYG000001443_03084 | |||||||||||
| CAZy Family | CBM2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 717245; End: 718405 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07995 | GSDH | 1.90e-67 | 64 | 370 | 1 | 326 | Glucose / Sorbosone dehydrogenase. Members of this family are glucose/sorbosone dehydrogenases that possess a beta-propeller fold. |
| COG2133 | YliI | 4.15e-53 | 17 | 309 | 20 | 325 | Glucose/arabinose dehydrogenase, beta-propeller fold [Carbohydrate transport and metabolism]. |
| TIGR03606 | non_repeat_PQQ | 7.37e-39 | 58 | 313 | 23 | 314 | dehydrogenase, PQQ-dependent, s-GDH family. PQQ, or pyrroloquinoline-quinone, serves as a cofactor for a number of sugar and alcohol dehydrogenases in a limited number of bacterial species. Most characterized PQQ-dependent enzymes have multiple repeats of a sequence region described by pfam01011 (PQQ enzyme repeat), but this protein family in unusual in lacking that repeat. Below the noise cutoff are related proteins mostly from species that lack PQQ biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCB78721.1 | 5.14e-109 | 58 | 383 | 47 | 360 |
| BCB83593.1 | 8.18e-109 | 64 | 383 | 53 | 360 |
| AVT28600.1 | 2.27e-105 | 58 | 383 | 49 | 361 |
| AVT38160.1 | 2.27e-105 | 58 | 383 | 49 | 361 |
| BCJ60637.1 | 4.99e-104 | 7 | 386 | 5 | 386 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DAS_A | 4.80e-171 | 43 | 385 | 10 | 346 | Structureof the PQQ-bound form of Aldose Sugar Dehydrogenase (Adh) from Streptomyces coelicolor [Streptomyces coelicolor] |
| 3A9G_A | 5.02e-64 | 57 | 383 | 21 | 348 | CrystalStructure of PQQ-dependent sugar dehydrogenase apo-form [Pyrobaculum aerophilum],3A9H_A Crystal Structure of PQQ-dependent sugar dehydrogenase holo-form [Pyrobaculum aerophilum] |
| 2ISM_A | 1.87e-63 | 50 | 383 | 16 | 351 | Crystalstructure of the putative oxidoreductase (glucose dehydrogenase) (TTHA0570) from thermus theromophilus HB8 [Thermus thermophilus HB8],2ISM_B Crystal structure of the putative oxidoreductase (glucose dehydrogenase) (TTHA0570) from thermus theromophilus HB8 [Thermus thermophilus HB8] |
| 7CDY_A | 7.53e-28 | 57 | 370 | 4 | 340 | ChainA, glucose dehydrogenase [Serratia sp. FS14],7CDY_B Chain B, glucose dehydrogenase [Serratia sp. FS14],7CGZ_A Chain A, glucose dehydrogenase [Serratia sp. FS14],7CGZ_B Chain B, glucose dehydrogenase [Serratia sp. FS14] |
| 5MIN_A | 2.05e-25 | 58 | 279 | 20 | 270 | ChainA, Quinoprotein glucose dehydrogenase B [Acinetobacter calcoaceticus],5MIN_B Chain B, Quinoprotein glucose dehydrogenase B [Acinetobacter calcoaceticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P73001 | 9.20e-27 | 64 | 308 | 65 | 342 | Uncharacterized protein slr1608 OS=Synechocystis sp. (strain PCC 6803 / Kazusa) OX=1111708 GN=slr1608 PE=3 SV=1 |
| P13650 | 3.45e-24 | 58 | 279 | 44 | 294 | Quinoprotein glucose dehydrogenase B OS=Acinetobacter calcoaceticus OX=471 GN=gdhB PE=1 SV=1 |
| P75804 | 1.15e-21 | 57 | 277 | 27 | 255 | Aldose sugar dehydrogenase YliI OS=Escherichia coli (strain K12) OX=83333 GN=yliI PE=1 SV=1 |
| Q96JK4 | 2.60e-14 | 110 | 279 | 246 | 444 | HHIP-like protein 1 OS=Homo sapiens OX=9606 GN=HHIPL1 PE=1 SV=2 |
| Q14DK5 | 3.49e-14 | 110 | 279 | 252 | 450 | HHIP-like protein 1 OS=Mus musculus OX=10090 GN=Hhipl1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000065 | 0.000000 | 0.000000 | 0.000000 |