You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001429_00848
You are here: Home > Sequence: MGYG000001429_00848
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
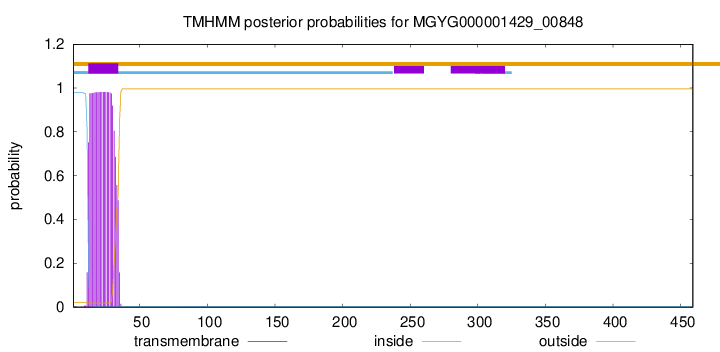
TMHMM annotations
Basic Information help
| Species | Alistipes_A ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A ihumii | |||||||||||
| CAZyme ID | MGYG000001429_00848 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 180802; End: 182181 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 65 | 252 | 6.9e-21 | 0.45614035087719296 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 2.26e-35 | 63 | 423 | 1 | 338 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 4.74e-12 | 66 | 176 | 1 | 100 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| pfam02894 | GFO_IDH_MocA_C | 4.40e-11 | 212 | 424 | 2 | 203 | Oxidoreductase family, C-terminal alpha/beta domain. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| pfam03435 | Sacchrp_dh_NADP | 5.84e-04 | 70 | 164 | 3 | 94 | Saccharopine dehydrogenase NADP binding domain. This family contains the NADP binding domain of saccharopine dehydrogenase. In some organisms this enzyme is found as a bifunctional polypeptide with lysine ketoglutarate reductase. The saccharopine dehydrogenase can also function as a saccharopine reductase. |
| COG1748 | Lys9 | 0.001 | 70 | 164 | 6 | 94 | Saccharopine dehydrogenase, NADP-dependent [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23801.1 | 2.05e-283 | 2 | 459 | 3 | 453 |
| BCG53919.1 | 2.46e-256 | 2 | 459 | 3 | 459 |
| ABR45528.1 | 4.48e-226 | 2 | 456 | 3 | 454 |
| QRO18095.1 | 4.48e-226 | 2 | 456 | 3 | 454 |
| QRX63444.1 | 2.23e-225 | 2 | 459 | 3 | 456 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3EC7_A | 1.80e-07 | 63 | 221 | 21 | 168 | CrystalStructure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_B Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_C Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_D Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_E Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_F Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_G Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_H Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 4HKT_A | 2.16e-07 | 66 | 340 | 4 | 247 | Crystalstructure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_B Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_C Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_D Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021] |
| 3MOI_A | 3.33e-06 | 139 | 239 | 65 | 161 | Thecrystal structure of the putative dehydrogenase from Bordetella bronchiseptica RB50 [Bordetella bronchiseptica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q54728 | 3.34e-08 | 66 | 313 | 2 | 221 | Uncharacterized oxidoreductase SP_1686 OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_1686 PE=3 SV=2 |
| O68965 | 5.01e-08 | 66 | 340 | 3 | 246 | Inositol 2-dehydrogenase OS=Rhizobium meliloti (strain 1021) OX=266834 GN=idhA PE=1 SV=2 |
| Q9RK81 | 1.80e-07 | 5 | 226 | 6 | 214 | Glycosyl hydrolase family 109 protein OS=Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) OX=100226 GN=SCO0529 PE=3 SV=1 |
| Q7MWF4 | 4.16e-07 | 44 | 242 | 38 | 241 | Glycosyl hydrolase family 109 protein OS=Porphyromonas gingivalis (strain ATCC BAA-308 / W83) OX=242619 GN=PG_0664 PE=3 SV=2 |
| A9N564 | 6.83e-07 | 66 | 277 | 3 | 183 | Inositol 2-dehydrogenase OS=Salmonella paratyphi B (strain ATCC BAA-1250 / SPB7) OX=1016998 GN=iolG PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
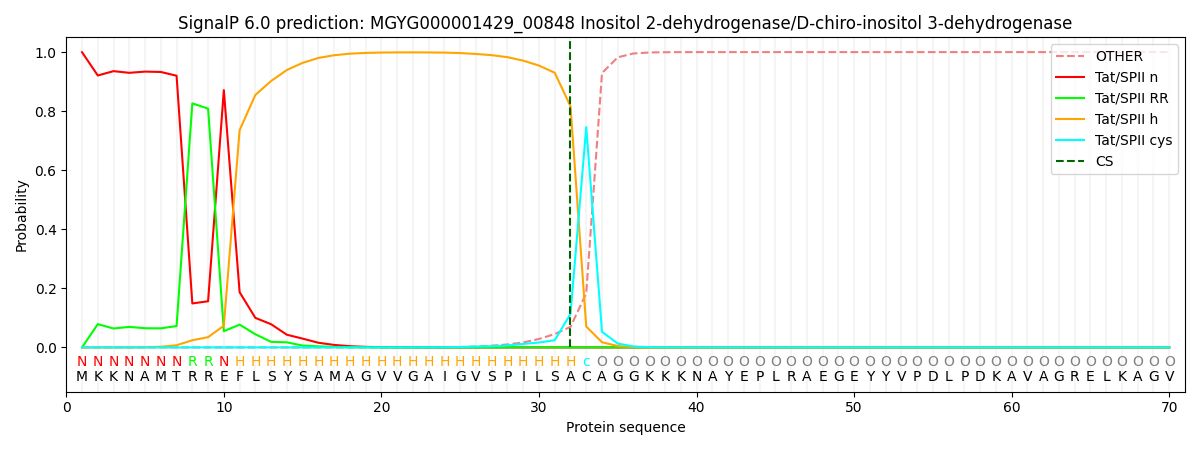
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000001 | 0.000348 | 0.999642 | 0.000000 |