You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001415_02197
You are here: Home > Sequence: MGYG000001415_02197
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes communis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes communis | |||||||||||
| CAZyme ID | MGYG000001415_02197 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 500226; End: 501716 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 99 | 396 | 4.8e-37 | 0.7122969837587007 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2723 | BglB | 2.01e-10 | 83 | 180 | 70 | 170 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| pfam01229 | Glyco_hydro_39 | 1.15e-09 | 101 | 435 | 70 | 420 | Glycosyl hydrolases family 39. |
| pfam00232 | Glyco_hydro_1 | 3.72e-06 | 56 | 211 | 30 | 216 | Glycosyl hydrolase family 1. |
| pfam02449 | Glyco_hydro_42 | 0.001 | 83 | 122 | 21 | 61 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| COG3693 | XynA | 0.002 | 73 | 179 | 47 | 151 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL03131.1 | 0.0 | 1 | 496 | 1 | 496 |
| BBL15167.1 | 0.0 | 1 | 496 | 1 | 496 |
| QNN22472.1 | 3.04e-157 | 1 | 474 | 1 | 467 |
| QNN22723.1 | 4.79e-106 | 41 | 477 | 35 | 473 |
| QBE67016.1 | 9.16e-101 | 35 | 477 | 18 | 482 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JVK_A | 3.82e-10 | 73 | 248 | 117 | 290 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
| 4EKJ_A | 3.18e-09 | 78 | 248 | 48 | 231 | ChainA, Beta-xylosidase [Caulobacter vibrioides] |
| 4M29_A | 3.18e-09 | 78 | 248 | 48 | 231 | Structureof a GH39 Beta-xylosidase from Caulobacter crescentus [Caulobacter vibrioides CB15] |
| 6YYH_A | 3.09e-08 | 105 | 257 | 100 | 262 | Crystalstructure of beta-D-xylosidase from Dictyoglomus thermophilum in ligand-free form [Dictyoglomus thermophilum H-6-12],6YYH_B Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum in ligand-free form [Dictyoglomus thermophilum H-6-12],6YYI_A Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose [Dictyoglomus thermophilum H-6-12],6YYI_B Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose [Dictyoglomus thermophilum H-6-12] |
| 1PX8_A | 2.12e-07 | 99 | 257 | 71 | 239 | Crystalstructure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1PX8_B Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_A Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_B Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_C Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum],1UHV_D Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase [Thermoanaerobacterium saccharolyticum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O30360 | 9.35e-08 | 105 | 257 | 77 | 239 | Beta-xylosidase OS=Thermoanaerobacterium saccharolyticum (strain DSM 8691 / JW/SL-YS485) OX=1094508 GN=xynB PE=3 SV=1 |
| P42973 | 3.67e-07 | 47 | 275 | 43 | 271 | Aryl-phospho-beta-D-glucosidase BglA OS=Bacillus subtilis (strain 168) OX=224308 GN=bglA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
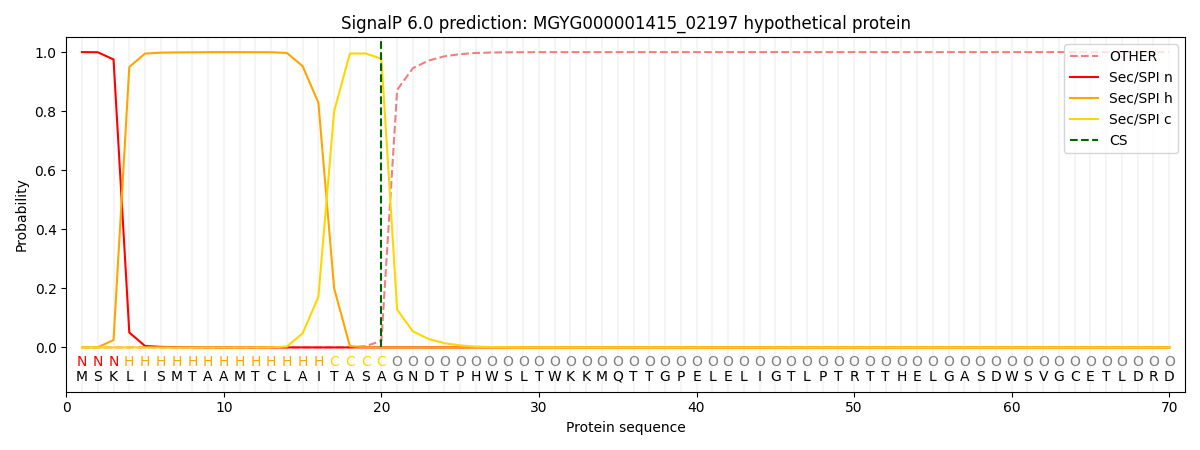
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000287 | 0.999079 | 0.000155 | 0.000166 | 0.000147 | 0.000138 |
