You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001412_03732
You are here: Home > Sequence: MGYG000001412_03732
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
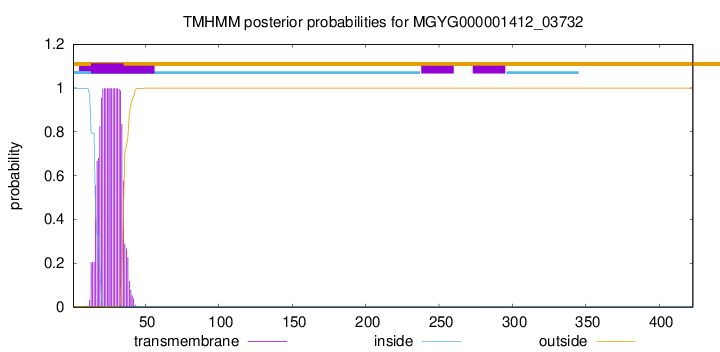
TMHMM annotations
Basic Information help
| Species | Robertmurraya massiliosenegalensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_B; DSM-18226; Robertmurraya; Robertmurraya massiliosenegalensis | |||||||||||
| CAZyme ID | MGYG000001412_03732 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 71872; End: 73143 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 81 | 374 | 3.2e-30 | 0.7331786542923434 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.75e-05 | 52 | 267 | 23 | 245 | Cellulase (glycosyl hydrolase family 5). |
| pfam01229 | Glyco_hydro_39 | 8.39e-04 | 100 | 284 | 86 | 295 | Glycosyl hydrolases family 39. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADF37974.1 | 1.02e-144 | 13 | 416 | 4 | 424 |
| QCR26351.1 | 3.83e-121 | 39 | 420 | 40 | 425 |
| QCR26354.1 | 2.34e-116 | 41 | 419 | 42 | 423 |
| AEN91003.1 | 5.66e-105 | 35 | 419 | 30 | 418 |
| ALC82660.1 | 1.46e-89 | 43 | 416 | 64 | 434 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JVK_A | 3.69e-22 | 41 | 395 | 101 | 473 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
| 4ZN2_A | 1.95e-09 | 46 | 290 | 21 | 286 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5BX9_A | 1.95e-09 | 46 | 290 | 21 | 286 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
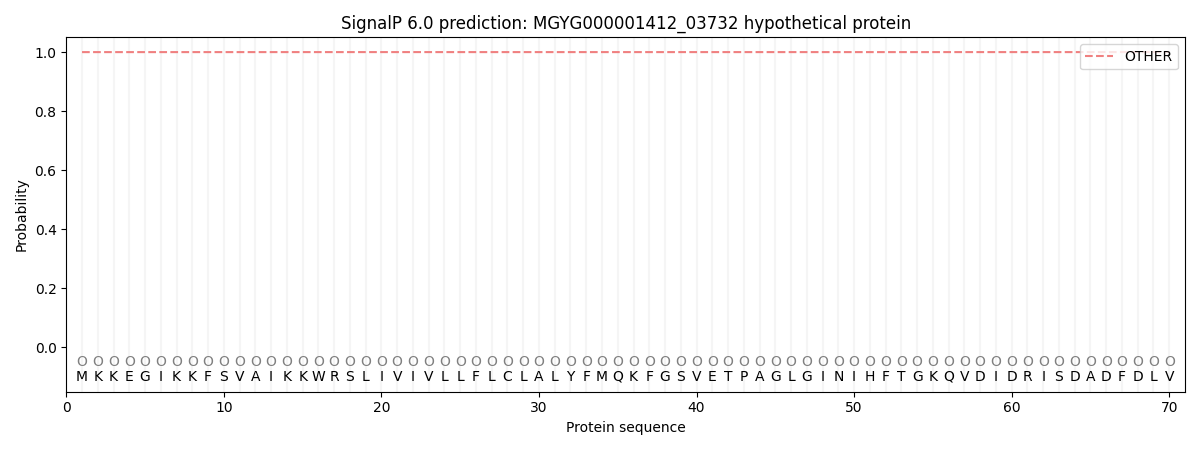
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999999 | 0.000019 | 0.000002 | 0.000000 | 0.000000 | 0.000001 |