You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001389_00571
You are here: Home > Sequence: MGYG000001389_00571
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cloacibacillus evryensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Synergistota; Synergistia; Synergistales; Synergistaceae; Cloacibacillus; Cloacibacillus evryensis | |||||||||||
| CAZyme ID | MGYG000001389_00571 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 99436; End: 101130 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 9 | 499 | 2.9e-132 | 0.9148148148148149 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13279 | arnT | 3.25e-81 | 7 | 559 | 3 | 551 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| COG1807 | ArnT | 4.04e-69 | 1 | 528 | 1 | 527 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 1.28e-17 | 66 | 225 | 2 | 159 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| pfam18583 | Arnt_C | 1.39e-06 | 461 | 518 | 21 | 66 | Aminoarabinose transferase C-terminal domain. ArnT is a member of the GT-C family of glycosyltransferases, and it has a similar fold to a bacterial oligosaccharyltransferase (OST) from Campylobacter lari (PglB) and to an archaeal OST from Archaeoglobus fulgidus (AglB). This entry represents the C-terminal periplasmic domain of Arnt proteins. |
| COG4745 | COG4745 | 0.002 | 3 | 179 | 9 | 177 | Predicted membrane-bound mannosyltransferase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANZ46074.1 | 0.0 | 1 | 564 | 1 | 556 |
| BDA10992.1 | 7.54e-132 | 4 | 556 | 15 | 558 |
| QIB60983.1 | 1.07e-131 | 4 | 556 | 15 | 558 |
| SNV04445.1 | 1.47e-131 | 18 | 556 | 19 | 546 |
| BDA10995.1 | 2.33e-130 | 18 | 556 | 19 | 546 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 1.14e-56 | 15 | 489 | 38 | 514 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2VBI7 | 1.40e-53 | 21 | 562 | 16 | 553 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Erwinia tasmaniensis (strain DSM 17950 / CFBP 7177 / CIP 109463 / NCPPB 4357 / Et1/99) OX=465817 GN=arnT PE=3 SV=1 |
| C5BDQ8 | 1.34e-52 | 15 | 562 | 10 | 552 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1 |
| A8FRR0 | 1.67e-52 | 20 | 558 | 17 | 541 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Shewanella sediminis (strain HAW-EB3) OX=425104 GN=arnT PE=3 SV=1 |
| A4WAM1 | 2.62e-52 | 21 | 562 | 16 | 553 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Enterobacter sp. (strain 638) OX=399742 GN=arnT PE=3 SV=1 |
| B4EUL1 | 2.33e-51 | 19 | 369 | 18 | 365 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Proteus mirabilis (strain HI4320) OX=529507 GN=arnT1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000036 | 0.000003 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
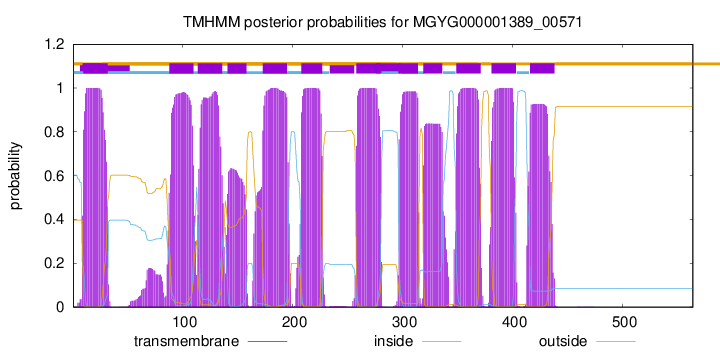
| start | end |
|---|---|
| 10 | 32 |
| 88 | 110 |
| 114 | 136 |
| 141 | 158 |
| 173 | 195 |
| 208 | 227 |
| 258 | 280 |
| 297 | 314 |
| 319 | 336 |
| 349 | 371 |
| 381 | 403 |
| 416 | 438 |
