You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001377_01365
You are here: Home > Sequence: MGYG000001377_01365
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
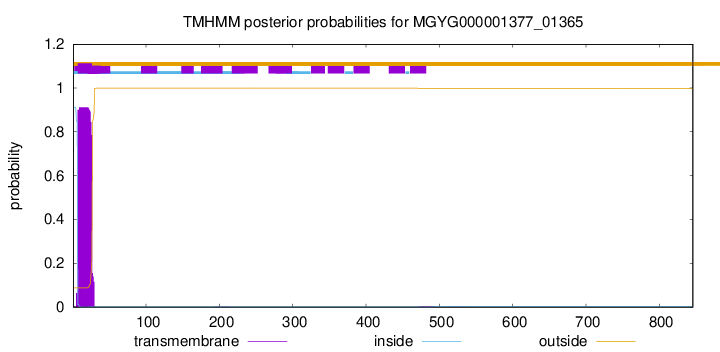
TMHMM annotations
Basic Information help
| Species | Dysgonomonas mossii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas mossii | |||||||||||
| CAZyme ID | MGYG000001377_01365 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 741048; End: 743588 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 137 | 603 | 5.3e-59 | 0.9904306220095693 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 2.67e-50 | 145 | 597 | 7 | 367 | Glycosyl hydrolase family 9. |
| cd10917 | CE4_NodB_like_6s_7s | 1.41e-25 | 637 | 836 | 1 | 170 | Catalytic NodB homology domain of rhizobial NodB-like proteins. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes many rhizobial NodB chitooligosaccharide N-deacetylase (EC 3.5.1.-)-like proteins, mainly from bacteria and eukaryotes, such as chitin deacetylases (EC 3.5.1.41), bacterial peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.-), and acetylxylan esterases (EC 3.1.1.72), which catalyze the N- or O-deacetylation of substrates such as acetylated chitin, peptidoglycan, and acetylated xylan. All members of this family contain a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold with 6- or 7 strands. Their catalytic activity is dependent on the presence of a divalent cation, preferably cobalt or zinc, and they employ a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. Several family members show diversity both in metal ion specificities and in the residues that coordinate the metal. |
| pfam02927 | CelD_N | 5.11e-25 | 24 | 127 | 1 | 83 | Cellulase N-terminal ig-like domain. |
| cd02850 | E_set_Cellulase_N | 6.25e-23 | 25 | 132 | 1 | 86 | N-terminal Early set domain associated with the catalytic domain of cellulase. E or "early" set domains are associated with the catalytic domain of cellulases at the N-terminal end. Cellulases are O-glycosyl hydrolases (GHs) that hydrolyze beta 1-4 glucosidic bonds in cellulose. They are usually categorized into either exoglucanases, which sequentially release terminal sugar units from the cellulose chain, or endoglucanases, which also attack the chain internally. The N-terminal domain of cellulase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
| cd10948 | CE4_BsPdaA_like | 4.97e-20 | 620 | 846 | 23 | 223 | Catalytic NodB homology domain of Bacillus subtilis polysaccharide deacetylase PdaA, and its bacterial homologs. The Bacillus subtilis genome contains six polysaccharide deacetylase gene homologs: pdaA, pdaB (previously known as ybaN), yheN, yjeA, yxkH and ylxY. This family is represented by Bacillus subtilis pdaA gene encoding polysaccharide deacetylase BsPdaA, which is a member of the carbohydrate esterase 4 (CE4) superfamily. BsPdaA deacetylates peptidoglycan N-acetylmuramic acid (MurNAc) residues to facilitate the formation of muramic delta-lactam, which is required for recognition of germination lytic enzymes. BsPdaA deficiency leads to the absence of muramic delta-lactam residues in the spore cortex. Like other CE4 esterases, BsPdaA consists of a single catalytic NodB homology domain that appears to adopt a deformed (beta/alpha)8 barrel fold with a putative substrate binding groove harboring the majority of the conserved residues. It utilizes a general acid/base catalytic mechanism involving a tetrahedral transition intermediate, where a water molecule functions as the nucleophile tightly associated to the zinc cofactor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIK60909.1 | 0.0 | 1 | 844 | 1 | 824 |
| QIK55492.1 | 0.0 | 1 | 845 | 1 | 825 |
| AGY53377.1 | 0.0 | 1 | 846 | 1 | 831 |
| QIA07887.1 | 2.53e-292 | 2 | 846 | 3 | 816 |
| AHW60805.1 | 1.97e-291 | 1 | 846 | 1 | 815 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3X17_A | 1.74e-31 | 22 | 602 | 14 | 551 | Crystalstructure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium],3X17_B Crystal structure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium] |
| 5U2O_A | 4.56e-25 | 75 | 605 | 20 | 537 | Crystalstructure of Zn-binding triple mutant of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
| 4CJ0_A | 1.72e-23 | 16 | 638 | 20 | 568 | ChainA, ENDOGLUCANASE D [Acetivibrio thermocellus],4CJ1_A Chain A, ENDOGLUCANASE D [Acetivibrio thermocellus] |
| 1CLC_A | 1.81e-23 | 16 | 638 | 34 | 582 | ChainA, ENDOGLUCANASE CELD; EC: 3.2.1.4 [Acetivibrio thermocellus] |
| 5U0H_A | 9.61e-22 | 75 | 605 | 20 | 537 | Crystalstructure of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C2S4 | 9.41e-23 | 16 | 638 | 20 | 568 | Endoglucanase D (Fragment) OS=Acetivibrio thermocellus OX=1515 GN=celD PE=1 SV=1 |
| A3DDN1 | 9.65e-22 | 16 | 638 | 44 | 592 | Endoglucanase D OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celD PE=1 SV=1 |
| P23658 | 8.01e-21 | 74 | 605 | 27 | 543 | Cellodextrinase OS=Butyrivibrio fibrisolvens OX=831 GN=ced1 PE=1 SV=1 |
| A7LXT3 | 1.21e-19 | 17 | 610 | 23 | 581 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH9A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02649 PE=1 SV=1 |
| A3DCH1 | 2.55e-18 | 26 | 610 | 214 | 814 | Cellulose 1,4-beta-cellobiosidase OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celK PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000211 | 0.999200 | 0.000174 | 0.000142 | 0.000134 | 0.000129 |