You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_01736
You are here: Home > Sequence: MGYG000001332_01736
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Massilioclostridium methylpentosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum | |||||||||||
| CAZyme ID | MGYG000001332_01736 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25126; End: 29097 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 41 | 975 | 5.4e-125 | 0.9575242718446602 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 1.31e-50 | 178 | 758 | 332 | 872 | alpha-L-rhamnosidase. |
| COG1566 | EmrA | 0.001 | 1168 | 1284 | 89 | 217 | Multidrug resistance efflux pump [Defense mechanisms]. |
| PRK04863 | mukB | 0.001 | 1005 | 1249 | 381 | 640 | chromosome partition protein MukB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AND79497.1 | 2.02e-124 | 12 | 990 | 11 | 995 |
| QDO03140.1 | 2.51e-115 | 10 | 996 | 16 | 997 |
| QDO34993.1 | 2.51e-115 | 10 | 996 | 16 | 997 |
| QDO45010.1 | 2.51e-115 | 10 | 996 | 16 | 997 |
| QDO24872.1 | 2.51e-115 | 10 | 996 | 16 | 997 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 2.26e-30 | 197 | 970 | 408 | 1104 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 6.52e-17 | 203 | 970 | 371 | 1063 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 1.47e-16 | 203 | 970 | 371 | 1063 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 1.21e-07 | 1140 | 1286 | 1667 | 1816 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
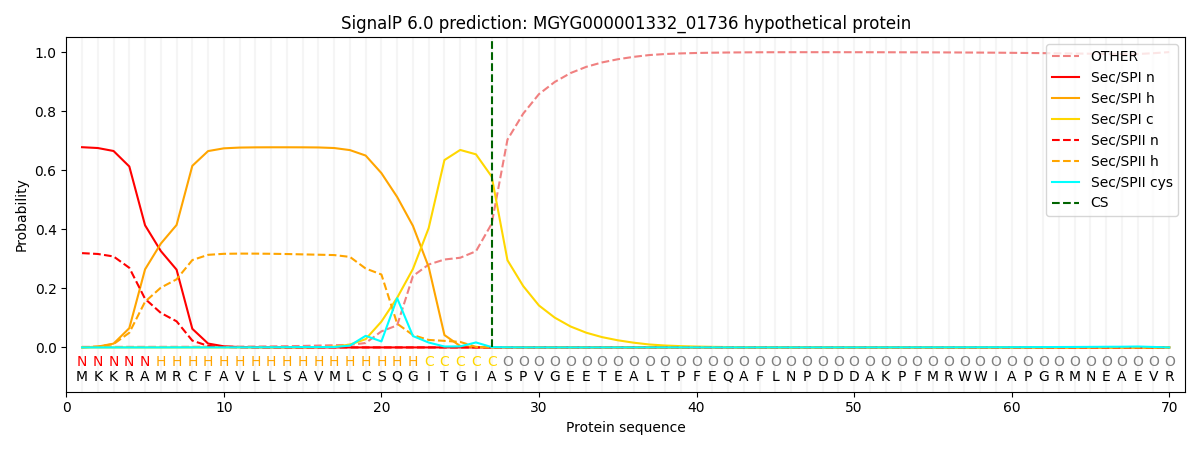
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.005923 | 0.657211 | 0.333624 | 0.001955 | 0.000716 | 0.000539 |

